Ubergraph Tutorial
Ubergraph is a SPARQL endpoint serving multiple OBO ontologies, pre-processed with:
relation-graph
information content scores
biolink categories
This notebook demonstrates the use of OAK with the ubergraph adapter. Many of the same operations can be applied with other adapters. Advantages of ubergraph include:
multiple ontologies all joined together in one graph
complex sparql querying of multiple pieces of information, including entailed relationships
First we set up a Jupyter alias for convenience:
[1]:
alias ubergraph runoak -i ubergraph:
First let’s see the list of ontologies in Ubergraph. Note this list is ever growing and if you try this you may see more than is shown here:
[2]:
ubergraph ontologies
aism/aism-base.owl
bspo.owl
chebi.owl
cl/cl-base.owl
cob/cob-base.owl
cob/components/cob-to-external.owl
colao/colao-base.owl
dpo/dpo-base.owl
eco/eco-base.owl
iao.owl
ecto/ecto-base.owl
emapa.owl
envo/envo-base.owl
fbbt/fbbt-base.owl
fbcv/fbcv-base.owl
fbdv/fbdv-base.owl
fypo/fypo-base.owl
go/extensions/go-bfo-bridge.owl
go/go-base.owl
hp/hp-base.owl
lepao/lepao-base.owl
ma.owl
maxo/maxo-base.owl
mi.owl
mmo.owl
mondo/mondo-base.owl
ncbitaxon.owl
ncit.owl
mp/mp-base.owl
nbo/nbo-base.owl
oba/oba-base.owl
obi/obi-base.owl
so.owl
pato/pato-base.owl
pcl/pcl-base.owl
pco/pco-base.owl
ro/ro-base.owl
uberon/bridge/cl-bridge-to-caro.owl
uberon/bridge/cl-bridge-to-fbbt.owl
uberon/bridge/uberon-bridge-to-caro.owl
uberon/bridge/uberon-bridge-to-fbbt.owl
uberon/uberon-base.owl
wbbt/wbbt-base.owl
wbls/wbls-base.owl
wbphenotype/wbphenotype-base.owl
zfa/zfa-base.owl
https://w3id.org/orcidio/orcidio.owl
pr/pr-asserted.owl
po.owl
https://raw.githubusercontent.com/PlantPhenoOntology/ppo/master/ppo.owl
apo.owl
mmusdv.owl
foodon.owl
to.owl
peco.owl
mro.owl
hao.owl
clao.owl
oarcs.owl
http://translator.renci.org/ubergraph-axioms.ofn
Basic lookup
We can use the OAK command to lookup various IDs
[24]:
ubergraph info CL:0000540 CL:0000679 RO:0002100 UBERON:0002771
CL:0000540 ! neuron
CL:0000679 ! glutamatergic neuron
WARNING:root:Multiple labels for RO:0002100 = has_soma_location != has soma location
WARNING:root:Multiple labels for RO:0002100 = has_soma_location != has soma location
RO:0002100 ! has_soma_location
UBERON:0002771 ! middle temporal gyrus
Note the annoying warning messages. OAK is telling us it couldn’t find the “definitive” label for RO:0002100. One challenge for a merged graph like Ubergraph is that some sources may include stale imports with older labels and metadata - worse, some may “inject” triples onto objects that don’t belong to them!
In future, OAK may include more advanced ways of retrieving labels such that trusted graphs are relied on over secondary ones; and it’s likely that some of these will be resolved in ubergraph.
But for now you can just use --quiet to silence the warnings.
Next let’s try querying for relationships:
[22]:
ubergraph --quiet relationships CL:0000540
subject subject_label predicate predicate_label object object_label
CL:0000540 neuron RO:0002215 capable of GO:0019226 transmission of nerve impulse
CL:0000540 neuron RO:0002216 capable of part of GO:0007154 cell communication
CL:0000540 neuron rdfs:subClassOf None CL:0000393 electrically responsive cell
CL:0000540 neuron rdfs:subClassOf None CL:0000404 electrically signaling cell
CL:0000540 neuron rdfs:subClassOf None CL:0002319 neural cell
By default, only asserted relationships are shown. Use --include-entailed to show also entailed relationships (calculated ahead of time using relation-graph):
[25]:
ubergraph --quiet relationships CL:0000540 -p p --include-entailed | head -30
Note that there a lot of trivial (but true) relationships there.
We can also use viz to visualize a subgraph, in this around “cell”, following is-a and part-of relationships:
[4]:
ubergraph viz -p i,p CL:0000540 -o output/ubergraph-neuron.png
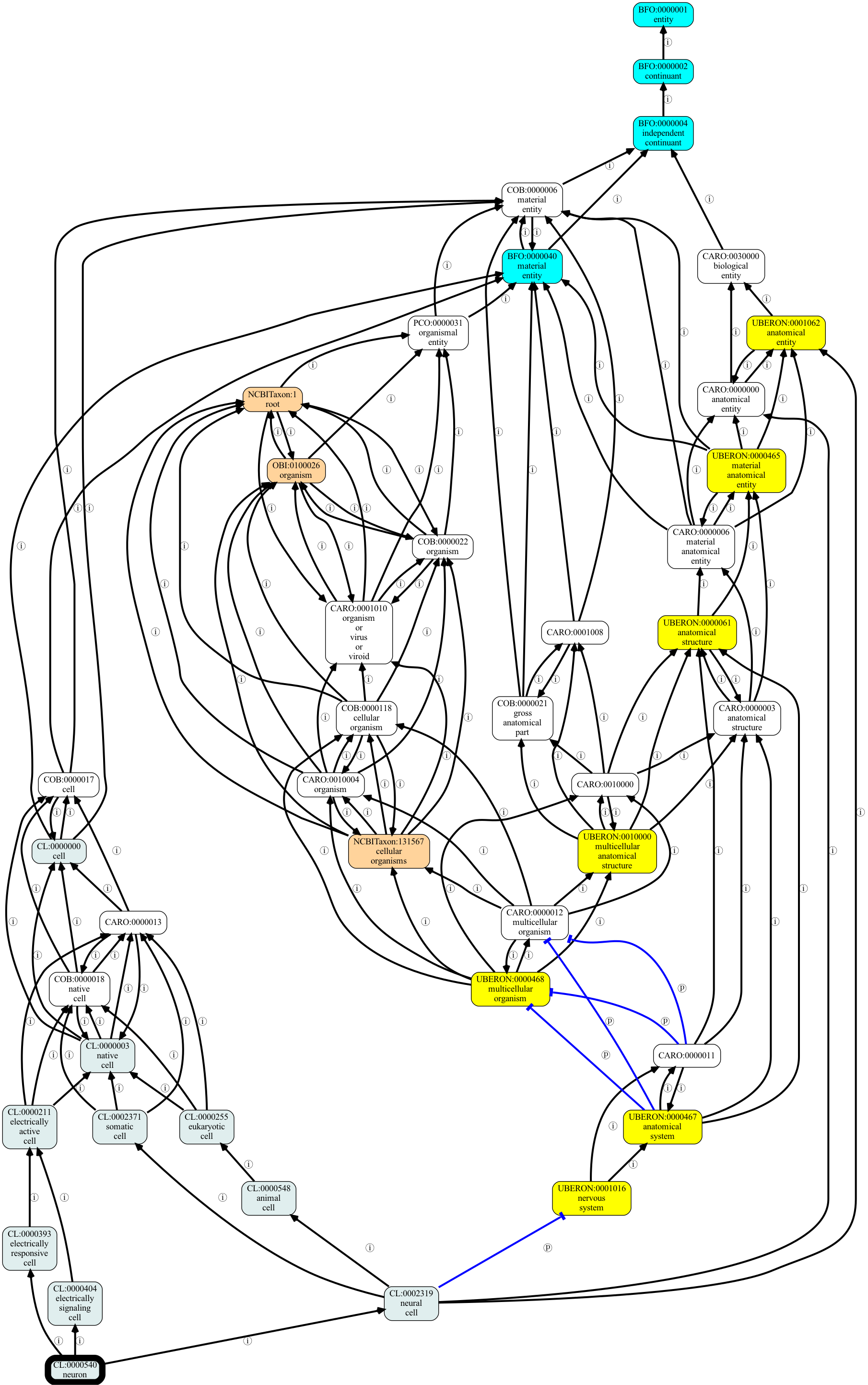
If you have used the viz command on individual ontologies before, you may notice a few differences with ubergraph.
Note that some pairs of terms have cyclic is-as pointing at one another: this is not an error! this reflects the fact that these two concepts are equivalent. In future we may add something at the visual layer that compacts these down into a bidirectional equivalence edge.
Making SPARQL queries
You can make SPARQL queries using the query command:
Here we will do a complex query that involves two relationship constraints:
all glutaminergic neurons (CL:0000679) that have
a soma location (RO:0002100) in the middle temporal gyrus (UBERON:0002771)
[16]:
ubergraph query -q "?s rdfs:subClassOf CL:0000679 ; RO:0002100 UBERON:0002771" -P CL,RO,UBERON
WARNING:root:Auto-adding limit
Query has no LIMIT clause: SELECT * WHERE {?s rdfs:subClassOf CL:0000679 ; RO:0002100 UBERON:0002771} LIMIT 100
s s_label
PCL:0023046 Exc L2 LAMP5 LTK middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023047 Exc L2-4 LINC00507 GLP2R middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023048 Exc L2-3 LINC00507 FREM3 middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023049 Exc L5-6 THEMIS C1QL3 middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023050 Exc L3-4 RORB CARM1P1 middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023051 Exc L3-5 RORB ESR1 middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023052 Exc L3-5 RORB COL22A1 middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023053 Exc L3-5 RORB FILIP1L middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023054 Exc L3-5 RORB TWIST2 middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023055 Exc L4-5 RORB FOLH1B middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023056 Exc L4-6 RORB SEMA3E middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023057 Exc L4-5 RORB DAPK2 middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023058 Exc L5-6 RORB TTC12 middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023059 Exc L4-6 RORB C1R middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023060 Exc L4-5 FEZF2 SCN4B middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023061 Exc L5-6 THEMIS DCSTAMP middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023062 Exc L5-6 THEMIS CRABP1 middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023063 Exc L5-6 THEMIS FGF10 middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023064 Exc L4-6 FEZF2 IL26 middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023065 Exc L5-6 FEZF2 ABO middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023066 Exc L6 FEZF2 SCUBE1 middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023067 Exc L5-6 FEZF2 IL15 middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023068 Exc L6 FEZF2 OR2T8 middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023069 Exc L5-6 FEZF2 EFTUD1P1 middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023127 L2/3 IT middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023130 L4 IT middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023134 L5 IT middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023138 L6 IT middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023142 L6b middle temporal gyrus glutamatergic neuron (Hsap)
Note that for your convenience, a surround SELECT {...} is added. The -P option will add prefix declarations for all specified prefixes using standard sources.
By default, labels will also be queried for all results (note that OAK will try to do this in an efficient a way as possible, avoiding iterative queries over the network). To suppress this, switch off the --autolabel option:
[17]:
ubergraph query -q "?s rdfs:subClassOf CL:0000679 ; RO:0002100 UBERON:0002771" -P CL,RO,UBERON --no-autolabel
WARNING:root:Auto-adding limit
Query has no LIMIT clause: SELECT * WHERE {?s rdfs:subClassOf CL:0000679 ; RO:0002100 UBERON:0002771} LIMIT 100
s
PCL:0023046
PCL:0023047
PCL:0023048
PCL:0023049
PCL:0023050
PCL:0023051
PCL:0023052
PCL:0023053
PCL:0023054
PCL:0023055
PCL:0023056
PCL:0023057
PCL:0023058
PCL:0023059
PCL:0023060
PCL:0023061
PCL:0023062
PCL:0023063
PCL:0023064
PCL:0023065
PCL:0023066
PCL:0023067
PCL:0023068
PCL:0023069
PCL:0023127
PCL:0023130
PCL:0023134
PCL:0023138
PCL:0023142
Advanced: feeding query results to other commands
So far we have seen an example of using the query command. You can actually use the results of a query in many other commands.
In OAK, most commands accept a query term list. These are typically lists of IDs or labels - but they can also be query expressions that are evaluated on the fly. One such expression is .query.
Here we will do the same query as above, and feed the results into the relationships command, allowing us the see the direct superclasses of all glutamine neurons in the MTG:
[30]:
ubergraph --quiet relationships -p i .query//prefixes=CL,RO,UBERON "?s rdfs:subClassOf CL:0000679 ; RO:0002100 UBERON:0002771"
subject subject_label predicate predicate_label object object_label
PCL:0023046 Exc L2 LAMP5 LTK middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None PCL:0023127 L2/3 IT middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023047 Exc L2-4 LINC00507 GLP2R middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None PCL:0023127 L2/3 IT middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023048 Exc L2-3 LINC00507 FREM3 middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None PCL:0023127 L2/3 IT middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023049 Exc L5-6 THEMIS C1QL3 middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None CL:0000679 glutamatergic neuron
PCL:0023049 Exc L5-6 THEMIS C1QL3 middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None CL:0010012 cerebral cortex neuron
PCL:0023050 Exc L3-4 RORB CARM1P1 middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None PCL:0023130 L4 IT middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023051 Exc L3-5 RORB ESR1 middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None PCL:0023130 L4 IT middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023052 Exc L3-5 RORB COL22A1 middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None PCL:0023130 L4 IT middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023053 Exc L3-5 RORB FILIP1L middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None PCL:0023130 L4 IT middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023054 Exc L3-5 RORB TWIST2 middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None PCL:0023130 L4 IT middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023055 Exc L4-5 RORB FOLH1B middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None PCL:0023134 L5 IT middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023056 Exc L4-6 RORB SEMA3E middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None PCL:0023134 L5 IT middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023057 Exc L4-5 RORB DAPK2 middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None PCL:0023134 L5 IT middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023058 Exc L5-6 RORB TTC12 middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None PCL:0023134 L5 IT middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023059 Exc L4-6 RORB C1R middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None PCL:0023134 L5 IT middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023060 Exc L4-5 FEZF2 SCN4B middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None CL:4023041 L5 extratelencephalic projecting glutamatergic cortical neuron
PCL:0023061 Exc L5-6 THEMIS DCSTAMP middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None PCL:0023138 L6 IT middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023062 Exc L5-6 THEMIS CRABP1 middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None PCL:0023138 L6 IT middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023063 Exc L5-6 THEMIS FGF10 middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None PCL:0023138 L6 IT middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023064 Exc L4-6 FEZF2 IL26 middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None CL:4023012 near-projecting glutamatergic cortical neuron
PCL:0023065 Exc L5-6 FEZF2 ABO middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None CL:4023042 L6 corticothalamic-projecting glutamatergic cortical neuron
PCL:0023066 Exc L6 FEZF2 SCUBE1 middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None PCL:0023142 L6b middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023067 Exc L5-6 FEZF2 IL15 middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None PCL:0023142 L6b middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023068 Exc L6 FEZF2 OR2T8 middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None PCL:0023142 L6b middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023069 Exc L5-6 FEZF2 EFTUD1P1 middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None PCL:0023142 L6b middle temporal gyrus glutamatergic neuron (Hsap)
PCL:0023127 L2/3 IT middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None CL:4023008 intratelencephalic-projecting glutamatergic cortical neuron
PCL:0023130 L4 IT middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None CL:4023008 intratelencephalic-projecting glutamatergic cortical neuron
PCL:0023134 L5 IT middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None CL:4023008 intratelencephalic-projecting glutamatergic cortical neuron
PCL:0023138 L6 IT middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None CL:4023008 intratelencephalic-projecting glutamatergic cortical neuron
PCL:0023142 L6b middle temporal gyrus glutamatergic neuron (Hsap) rdfs:subClassOf None CL:4023038 L6b glutamatergic cortical neuron
[ ]: