Association Provider Interface
- class oaklib.interfaces.association_provider_interface.AssociationProviderInterface(resource: ~oaklib.resource.OntologyResource = None, strict: bool = False, _multilingual: bool = None, autosave: bool = <factory>, exclude_owl_top_and_bottom: bool = <factory>, ontology_metamodel_mapper: ~oaklib.mappers.ontology_metadata_mapper.OntologyMetadataMapper | None = None, _converter: ~curies.api.Converter | None = None, auto_relax_axioms: bool = None, cache_lookups: bool = False, property_cache: ~oaklib.utilities.keyval_cache.KeyValCache = <factory>, _edge_index: ~oaklib.indexes.edge_index.EdgeIndex | None = None, _entailed_edge_index: ~oaklib.indexes.edge_index.EdgeIndex | None = None, _prefix_map: ~typing.Mapping[str, str] | None = None, _association_index: ~oaklib.utilities.associations.association_index.AssociationIndex = None, normalizers: ~typing.List[~oaklib.interfaces.association_provider_interface.EntityNormalizer] = <factory>)[source]
An ontology provider that provides associations.
Associations (also known as annotations) connect data elements or entities to an ontology class. Examples of associations include:
Gene associations to terms in ontologies like GO, Mondo, Uberon, HPO, MPO, CL
Associations between spans of text and ontology entities
Data models and file formats include:
The GO GAF and GPAD formats.
The HPOA association file format.
KGX (Knowledge Graph Exchange).
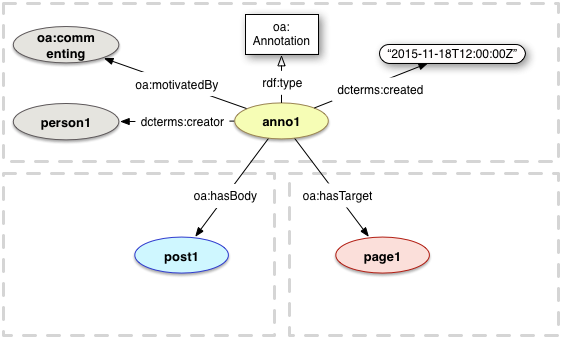
The W3 Open Annotation (OA) data model
The OA datamodel considers an annotation to be between a body and a target:
Warning
the signature of some methods are subject to change while we decide on the best patterns to define here.
Note that most ontology sources are not themselves providers of associations; there is no agreed upon way to represent associations in OWL, and associations are typically distributed separately from ontologies. See the section Associations and Curated Annotations in the OAK guide for more details.
OAK provides a number of ways to augment an ontology adapter with associations.
The most expressive way is to provide an InputSpecification:
>>> from oaklib import get_adapter >>> adapter = get_adapter("src/oaklib/conf/go-pombase-input-spec.yaml")
This combines an ontology source with one or more association sources.
Another approach is to use an adapter that directly supports associations. An example of this is the amigo_implementation:
>>> from oaklib import get_adapter >>> amigo = get_adapter("amigo:NCBITaxon:10090") # mouse
Command Line Use
runoak -i foo.db associations UBERON:0002101
- associations(subjects: Iterable[str] = None, predicates: Iterable[str] = None, objects: Iterable[str] = None, property_filter: Dict[str, Any] = None, subject_closure_predicates: List[str] | None = None, predicate_closure_predicates: List[str] | None = None, object_closure_predicates: List[str] | None = None, include_modified: bool = False, add_closure_fields: bool = False, **kwargs) Iterator[Association][source]
Yield all matching associations.
To query by subject (e.g. genes):
>>> from oaklib import get_adapter >>> adapter = get_adapter("src/oaklib/conf/go-pombase-input-spec.yaml") >>> genes = ["PomBase:SPAC1142.02c", "PomBase:SPAC3H1.05", "PomBase:SPAC1142.06", "PomBase:SPAC4G8.02c"] >>> for assoc in adapter.associations(genes): ... print(f"{assoc.object} {adapter.label(assoc.object)}") ... GO:0006620 post-translational protein targeting to endoplasmic reticulum membrane ...
To query by object (e.g. descriptor terms):
>>> from oaklib import get_adapter >>> from oaklib.datamodels.vocabulary import IS_A, PART_OF >>> adapter = get_adapter("src/oaklib/conf/go-pombase-input-spec.yaml") >>> for assoc in adapter.associations(objects=["GO:0006620"], object_closure_predicates=[IS_A, PART_OF]): ... print(f"{assoc.subject} {assoc.subject_label}") ... PomBase:SPAC1142.02c sgt2 ...
When determining a match on objects, the predicates in
object_closure_predicatesis used. We recommend you always explicitly provide this. A good choice is typically IS_A and PART_OF for ontologies like GO, Uberon, CL, ENVO.- Parameters:
subjects – constrain to these subjects (e.g. genes in a gene association)
predicates – constrain to these predicates (e.g. involved-in for a gene to pathway association)
objects – constrain to these objects (e.g. terms)
property_filter – generic query filter
subject_closure_predicates – subjects is treated as descendant via these predicates
predicate_closure_predicates – predicates is treated as descendant via these predicates
object_closure_predicates – object is treated as descendant via these predicates
add_closure_fields – add subject and object closure fields to the association
include_modified
- Returns:
- associations_subjects(*args, **kwargs) Iterator[str][source]
Yields all distinct subjects.
>>> from oaklib import get_adapter >>> from oaklib.datamodels.vocabulary import IS_A, PART_OF >>> adapter = get_adapter("src/oaklib/conf/go-pombase-input-spec.yaml") >>> preds = [IS_A, PART_OF] >>> for gene in adapter.associations_subjects(objects=["GO:0045047"], object_closure_predicates=preds): ... print(gene) ... PomBase:SPBC1271.05c ...
- Parameters:
kwargs – same arguments as for Associations and Curated Annotations
- Returns:
- associations_subject_search(subjects: Iterable[str] = None, predicates: Iterable[str] = None, objects: Iterable[str] = None, property_filter: Dict[str, Any] = None, subject_closure_predicates: List[str] | None = None, predicate_closure_predicates: List[str] | None = None, object_closure_predicates: List[str] | None = None, subject_prefixes: List[str] | None = None, include_similarity_object: bool = False, method: str | None = None, limit: int | None = 10, sort_by_similarity: bool = True, **kwargs) Iterator[Tuple[float, TermSetPairwiseSimilarity | None, str]][source]
Search over all subjects in the association index.
This relies on the SemanticSimilarityInterface.
Note
this is currently quite slow, this will be optimized in future
- Parameters:
subjects – optional set of subjects (e.g. genes) to search against
predicates – only use associations with this predicate
objects – this is the query - the asserted objects for all subjects
property_filter – passed to associations query
subject_closure_predicates – passed to associations query
predicate_closure_predicates – passed to associations query
object_closure_predicates – closure to use over the ontology
subject_prefixes – only consider subjects with these prefixes
include_similarity_object – include the similarity object in the result
method – similarity method to use
limit – max number of results to return
kwargs
- Returns:
iterator over ordered pairs of (score, sim, subject)
- association_pairwise_coassociations(curies1: Iterable[str], curies2: Iterable[str], inputs_are_subjects=False, include_reciprocals=False, include_diagonal=True, include_entities=True, include_negated=False, **kwargs) Iterator[PairwiseCoAssociation][source]
Find co-associations.
>>> from oaklib import get_adapter >>> from oaklib.datamodels.vocabulary import IS_A, PART_OF >>> adapter = get_adapter("src/oaklib/conf/go-pombase-input-spec.yaml") >>> terms = ["GO:0000910", "GO:0006281", "GO:0006412"] >>> preds = [IS_A, PART_OF] >>> for coassoc in adapter.association_pairwise_coassociations(curies1=terms, ... curies2=terms, ... object_closure_predicates=preds): ... print(coassoc.object1, coassoc.object2, coassoc.number_subjects_in_common) ... GO:0006281 GO:0000910 0 ...
- Parameters:
curies1
curies2
inputs_are_subjects
kwargs
- Returns:
- add_associations(associations: Iterable[Association], normalizers: List[EntityNormalizer] = None, **kwargs) bool[source]
Store a collection of associations for later retrievals.
- Parameters:
associations
normalizers
- Returns:
- association_counts(subjects: Iterable[str] = None, predicates: Iterable[str] = None, property_filter: Dict[str, Any] = None, subject_closure_predicates: List[str] | None = None, predicate_closure_predicates: List[str] | None = None, object_closure_predicates: List[str] | None = None, include_modified: bool = False, group_by: str | None = 'object', limit: int | None = None, **kwargs) Iterator[Tuple[str, int]][source]
Yield objects together with the number of distinct associations.
- Parameters:
subjects
predicates
property_filter
subject_closure_predicates
predicate_closure_predicates
object_closure_predicates
include_modified
group_by
limit
kwargs
- Returns:
- association_subject_counts(subjects: Iterable[str] = None, predicates: Iterable[str] = None, property_filter: Dict[str, Any] = None, subject_closure_predicates: List[str] | None = None, predicate_closure_predicates: List[str] | None = None, object_closure_predicates: List[str] | None = None, include_modified: bool = False, **kwargs) Iterator[Tuple[str, int]][source]
Yield objects together with the number of distinct associated subjects.
Here objects are typically nodes from ontologies and subjects are annotated entities such as genes.
>>> from oaklib import get_adapter >>> from oaklib.datamodels.vocabulary import IS_A, PART_OF >>> adapter = get_adapter("src/oaklib/conf/go-pombase-input-spec.yaml") >>> genes = ["PomBase:SPAC1142.02c", "PomBase:SPAC3H1.05", "PomBase:SPAC1142.06"] >>> preds = [IS_A, PART_OF] >>> for term, num in adapter.association_subject_counts(genes, object_closure_predicates=preds): ... print(term, num) ... GO:0051668 3 ...
This shows that GO:0051668 (localization within membrane) is used for all 3 input subjects. If subjects is empty, this is calculated for all subjects in the association set.
- Parameters:
subjects – constrain to these subjects (e.g. genes in a gene association)
predicates – constrain to these predicates (e.g. involved-in for a gene to pathway association)
property_filter – generic filter
subject_closure_predicates – subjects is treated as descendant via these predicates
predicate_closure_predicates – predicates is treated as descendant via these predicates
object_closure_predicates – object is treated as descendant via these predicates
include_modified – include modified associations
kwargs – additional arguments
- Returns:
- map_associations(subjects: Iterable[str] = None, predicates: Iterable[str] = None, objects: Iterable[str] = None, subset: str = None, subset_entities: Iterable[str] = None, property_filter: Dict[str, Any] = None, subject_closure_predicates: List[str] | None = None, predicate_closure_predicates: List[str] | None = None, object_closure_predicates: List[str] | None = None, include_modified: bool = False) Iterator[Association][source]
Maps matching associations to a subset (map2slim, rollup).
- Parameters:
subjects – constrain to these subjects
predicates – constrain to these predicates (e.g. involved-in for a gene to pathway association)
objects – constrain to these objects (e.g. terms)
subset – subset to map to
subset_entities – subset entities to map to
property_filter – generic filter
subject_closure_predicates – subjects is treated as descendant via these predicates
predicate_closure_predicates – predicates is treated as descendant via these predicates
object_closure_predicates – object is treated as descendant via these predicates
include_modified – include modified associations
- Returns: