OAK apply command
This notebook is intended as a supplement to the main OAK CLI docs.
This notebook provides examples for the apply command, which applies any change conforming to the KGCL specification.
Help Option
You can get help on any OAK command using --help
[1]:
!runoak apply --help
Usage: runoak apply [OPTIONS] [COMMANDS]...
Applies a patch to an ontology. The patch should be specified using KGCL
syntax, see https://github.com/INCATools/kgcl
Example:
runoak -i cl.owl.ttl apply "rename CL:0000561 to 'amacrine neuron'" -o
cl.owl.ttl -O ttl
On an obo format file:
runoak -i simpleobo:go-edit.obo apply "rename GO:0005634 from 'nucleus'
to 'foo'" -o go-edit-new.obo
With URIs:
runoak -i cl.owl.ttl apply "rename
<http://purl.obolibrary.org/obo/CL_0000561> from 'amacrine cell' to
'amacrine neuron'" -o cl.owl.ttl -O ttl
WARNING:
This command is still experimental. Some things to bear in mind:
- for some ontologies, CURIEs may not work, instead specify a full URI
surrounded by <>s - only a subset of KGCL commands are supported by each
backend
Options:
-o, --output TEXT
--changes-output TEXT output file for KGCL changes
--changes-input FILENAME Path to an input changes file
--changes-format TEXT Format of the changes file (json or kgcl)
--dry-run / --no-dry-run if true, only perform the parse of KCGL and
do not apply [default: no-dry-run]
--expand / --no-expand if true, expand complex changes to atomic
changes [default: expand]
--ignore-invalid-changes / --no-ignore-invalid-changes
if true, ignore invalid changes, e.g.
obsoletions of dependent entities [default:
no-ignore-invalid-changes]
--contributor TEXT CURIE for the person contributing the patch
-O, --output-type TEXT Desired output type
--overwrite / --no-overwrite If set, any changes applied will be saved
back to the input file/source
--help Show this message and exit.
Download example file
A typical use case for the apply command is for applying changes to the source, aka edit version of an ontology. For our purposes here we will make a copy of the go editorial file.
[7]:
!curl -L -s https://github.com/geneontology/go-ontology/raw/master/src/ontology/go-edit.obo > input/go-edit.obo
Note that the go edit file is in obo format. A number of ontologies like GO, Uberon, and Mondo use obo format as the edit format due to the fact obo was designed to make human-readable diffs.
The KGCL apply command may be used with other adapters, but it has been tested most extensively on the above three ontologies.
Create a new exact synonym
Next we will create a new change of type NewSynonym, using KGCL syntax on the command line.
We will try making a synonym compartment for GO:0043226 (organelle)
We will first run in --dry-run mode:
[3]:
!runoak -i simpleobo:input/go-edit.obo apply "create exact synonym 'compartment' for GO:0043226" --dry-run
WARNING:root:--autosave not passed, changes are NOT saved
create exact synonym 'compartment' for GO:0043226
This warns us that changes were not saved anywhere.
next we will try the real deal, and save the output file:
[4]:
!runoak -i simpleobo:input/go-edit.obo apply "create exact synonym 'compartment' for GO:0043226" -o output/go-edit-modified.obo
The command doesn’t produce any output on stdout, but we instructed it to save these in an external file output/go-edit-modified.obo.
Let’s double check that it did what we asked it to do. First we’ll try a plain old unix diff (one advantage of OBO format is its easy diffability):
[5]:
!diff -u input/go-edit.obo output/go-edit-modified.obo
--- input/go-edit.obo 2023-01-20 12:36:57.000000000 -0800
+++ output/go-edit-modified.obo 2023-01-20 12:37:07.000000000 -0800
@@ -241846,6 +241846,7 @@
xref: NIF_Subcellular:sao1539965131
xref: Wikipedia:Organelle
is_a: GO:0110165 ! cellular anatomical entity
+synonym: "compartment" EXACT []
[Term]
id: GO:0043227
This is also what you would see in a Pull Request implementing this change
Diff Command
The unix diff is still a little low level. OAK comes with a diff command that we can use instead.
This is the reciprocal of the apply command, and it will generate a set of change objects in KGCL (which can then be applied….)
[5]:
!runoak -i simpleobo:input/go-edit.obo diff -X simpleobo:output/go-edit-modified.obo -O json
[
{
"id": "uuid:a50afe2c-9ed4-4ee9-9a17-e80e971b072e",
"new_value": "compartment",
"about_node": "GO:0043226",
"@type": "NewSynonym"
}
]
(this is currently a bit slow, so be patient - we’re working on optimizing this).
If you prefer human-readable KGCL syntax to KGCL JSON:
[6]:
!runoak -i simpleobo:input/go-edit.obo diff -X simpleobo:output/go-edit-modified.obo -O kgcl
create synonym 'compartment' for GO:0043226
Note that this is the same string we used to apply the patch in the first place - this demonstrates the complementary nature of diff and patch.
TODO: the diff should reflect the scope of the synonym, i.e EXACT
Apply multiple changes
You can pass in a list of multiple changes on the command line, or via a file:
[11]:
!echo create exact synonym \'test1\' for GO:0043226 > input/test.kgcl
[12]:
!echo create exact synonym \'test2\' for GO:0043226 >> input/test.kgcl
[13]:
!cat input/test.kgcl
create exact synonym 'test1' for GO:0043226
create exact synonym 'test2' for GO:0043226
[14]:
!runoak -i simpleobo:input/go-edit.obo apply --changes-input input/test.kgcl -o output/go-edit-modified.obo
Expanding complex changes into atomic changes
Some changes represent composites of multiple smaller changes; other changes might entail other changes. Some of these may be variable depending on particular ontology workflows.
For example, in many OBO workflows, the act of performing a NodeObsoletion might also involve:
renaming the node, preceding the label with “
obsolete”rewiring the surrounding nodes, such that:
the children of the obsolete nodes point directly to the parents, with the obsolete node bypassed
deleting edges such that there are no logical axioms that reference the obsoleted node
first let’s try a dry run simulating what it would be like to obsolete organelle (GO:0043226).
First let’s explore the neighborhood - we will use the viz command to view a random child of organelle, non-membrane-bounded organelle (GO:0043228)
[23]:
!runoak -i simpleobo:input/go-edit.obo viz -p i,p GO:0043228 GO:0043226 -o output/nmbo.png
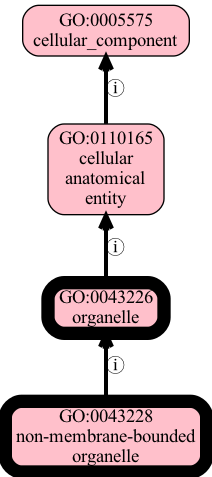
now let’s try obsoleting the intermediate organelle class (GO:0043226), but in --dry-run mode, with --expand. (Note --expand is the default, but it helps to make this explicit).
This will trigger the outputting of all expanded changes as KGCL syntax:
[17]:
!runoak -i simpleobo:input/go-edit.obo apply --expand "obsolete GO:0043226" --dry-run
obsolete GO:0043226
rename GO:0043226 from 'organelle' to 'obsolete organelle'
create edge GO:0005929 rdfs:subClassOf GO:0110165
create edge GO:0043228 rdfs:subClassOf GO:0110165
create edge GO:0043227 rdfs:subClassOf GO:0110165
create edge GO:0043230 rdfs:subClassOf GO:0110165
create edge GO:0099572 rdfs:subClassOf GO:0110165
delete edge GO:0005929 rdfs:subClassOf GO:0043226
delete edge GO:0043228 rdfs:subClassOf GO:0043226
delete edge GO:0020004 BFO:0000050 GO:0043226
delete edge GO:0031676 BFO:0000050 GO:0043226
delete edge GO:0043227 rdfs:subClassOf GO:0043226
delete edge GO:0032420 BFO:0000050 GO:0043226
delete edge GO:0043230 rdfs:subClassOf GO:0043226
delete edge GO:0044232 BFO:0000050 GO:0043226
delete edge GO:0060091 BFO:0000050 GO:0043226
delete edge GO:0060171 BFO:0000050 GO:0043226
delete edge GO:0097591 BFO:0000050 GO:0043226
delete edge GO:0097592 BFO:0000050 GO:0043226
delete edge GO:0097593 BFO:0000050 GO:0043226
delete edge GO:0097594 BFO:0000050 GO:0043226
delete edge GO:0097595 BFO:0000050 GO:0043226
delete edge GO:0097596 BFO:0000050 GO:0043226
delete edge GO:0099572 rdfs:subClassOf GO:0043226
delete edge GO:0043226 rdfs:subClassOf GO:0110165WARNING:root:--autosave not passed, changes are NOT saved
in future it will be possible to visualize KGCL directly. For now, let’s just visualize the output file after running in non-dry-run mode:
[19]:
!runoak -i simpleobo:input/go-edit.obo apply --expand "obsolete GO:0043226" -o output/obsoleted-organelle.obo
[22]:
!runoak --stacktrace -i simpleobo:output/obsoleted-organelle.obo viz -p i,p GO:0043228 GO:0043226 -o output/nmbo2.png
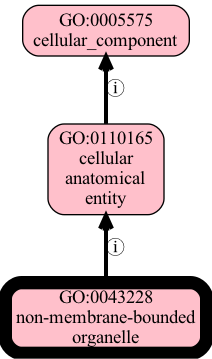
Invalid Obsolete Operations
Currently the obsolete operation will not rewire certain axioms of ontology axioms like logical definitions, these require curator intervention.
This can be seen if we try and obsolete a core term like metabolic process (GO:0008152):
[15]:
!runoak -i simpleobo:input/go-edit.obo apply --expand "obsolete GO:0008152" --dry-run
ValueError: GO:0008152 used in logical definition of GO:0000023
In future, OAK may allow more configurability here, including the ability to do full cascading deletes. But this in general would not be recommended - if you want to obsolete a term that is commonly used in logical definitions then you need to do some manual examination of your design patterns.
However, if you also want to obsolete all the dependent nodes in the same operation, you can do that by batching the obsoletes in a single file.
Creating an entire ontology from change directives
You can create an entire ontology from scratch using only change directives.
[17]:
!cat input/test-create.kgcl.txt
create node X:1 'limb'
create node X:2 'forelimb'
create edge X:2 is_a X:1
create node X:3 'hindlimb'
create edge X:3 is_a X:1
create related synonym 'arm' for X:2
create related synonym 'leg' for X:3
# foo
[19]:
!runoak -i pronto: apply --changes-input input/test-create.kgcl.txt -o output/kgcl-de-novo.obo
[20]:
!cat output/kgcl-de-novo.obo
format-version: 1.4
[Term]
id: X:1
name: limb
[Term]
id: X:2
name: forelimb
synonym: "arm" RELATED []
is_a: X:1
[Term]
id: X:3
name: hindlimb
synonym: "leg" RELATED []
is_a: X:1
the same thing but using the funowl wrapper for making an ontology in OWL functional syntax. Note here it’s necessary to set the prefixes as these are not implicit like in obo:
[1]:
!runoak --stacktrace --prefix X=http://example.org/ -i funowl: apply --changes-input input/test-create.kgcl.txt -o output/kgcl-de-novo.ofn
[2]:
!cat output/kgcl-de-novo.ofn
Prefix( owl: = <http://www.w3.org/2002/07/owl#> )
Prefix( rdf: = <http://www.w3.org/1999/02/22-rdf-syntax-ns#> )
Prefix( rdfs: = <http://www.w3.org/2000/01/rdf-schema#> )
Prefix( xsd: = <http://www.w3.org/2001/XMLSchema#> )
Prefix( xml: = <http://www.w3.org/XML/1998/namespace> )
Ontology(
AnnotationAssertion( rdfs:label <http://example.org/1> "limb" )
AnnotationAssertion( rdfs:label <http://example.org/2> "forelimb" )
SubClassOf( <http://example.org/2> <http://example.org/1> )
AnnotationAssertion( rdfs:label <http://example.org/3> "hindlimb" )
SubClassOf( <http://example.org/3> <http://example.org/1> )
AnnotationAssertion( <http://www.geneontology.org/formats/oboInOwl#hasExactSynonym> X:2 "arm" )
AnnotationAssertion( <http://www.geneontology.org/formats/oboInOwl#hasExactSynonym> X:3 "leg" )
)
[ ]: