OAK logical-definitions command
This notebook is intended as a supplement to the main OAK CLI docs.
This notebook provides examples for the logical-definitions command, which can be used to lookup and summarize logical defs
For more on logical definitions, see Logical Definitions in the OAK guide.
Help Option
You can get help on any OAK command using --help
[1]:
!runoak logical-definitions --help
Usage: runoak logical-definitions [OPTIONS] [TERMS]...
Show all logical definitions for a term or terms.
To show all logical definitions in an ontology, pass the ".all" query term
Example; first create an alias:
alias pato="runoak -i obo:sqlite:pato"
Then run the query:
pato logical-definitions .all
By default, ".all" will query all axioms for all terms including merged
terms; to restrict to only the current terms, use an ID query:
pato logical-definitions i^PATO
You can also restrict to branches:
pato logical-definitions .desc//p=i "physical object quality"
By default, the output is a subset of OboGraph datamodel rendered as YAML,
e.g.
definedClassId: PATO:0045071 genusIds: - PATO:0001439
restrictions: - fillerId: PATO:0000461 propertyId: RO:0015010
You can also specify CSV to generate a flattened form of this.
Example:
pato logical-definitions .all --output-type csv
You can optionally choose to "--matrix-axes" to transform the output to a
matrix form. This is a comma-separated pair of axes, where each element is a
logical definition element type: "f" for filler, "p" for predicate, "g" for
genus, "d" for defined class.
Example:
- Each property/predicate is a column - For repeated properties, columns of
the form prop_1, prop_2, ... are generated
Example:
pato logical-definitions .all --matrix-axes d,p --output-type csv
This will generate a row for each defined class with a logical definition,
with columns for each predicate ("genus" is treated as a predicate here).
Limitations:
Currently this only works for definitions that follow a basic genus-
differentia pattern, which is what is currently represented in the OboGraph
datamodel.
Consider using the "axioms" command for inspection of complex nested OWL
axioms.
More examples:
https://github.com/INCATools/ontology-access-
kit/blob/main/notebooks/Commands/LogicalDefinitions.ipynb
Python API:
https://incatools.github.io/ontology-access-kit/interfaces/obograph
Data model:
https://w3id.org/oak/obograph
Options:
--unmelt / --no-unmelt Flatten to a wide table [default: no-
unmelt]
--matrix-axes TEXT If specified, transform results to matrix
using these row and column axes. Examples:
d,p; f,g
-p, --predicates TEXT A comma-separated list of predicates. This
may be a shorthand (i, p) or CURIE
--autolabel / --no-autolabel If set, results will automatically have
labels assigned [default: autolabel]
-O, --output-type TEXT Desired output type
-o, --output FILENAME Output file, e.g. obo file
--if-absent [absent-only|present-only]
determines behavior when the value is not
present or is empty.
-S, --set-value TEXT the value to set for all terms for the given
property.
--help Show this message and exit.
Set up an alias
For convenience we will set up an alias for use in this notebook
[2]:
alias uberon runoak -i sqlite:obo:uberon
Fetching logical definitions for individual terms
First we will pass in a simple list of terms to the command.
Like most OAK commands, this command accepts lists of either IDs, labels, queries, or boolean combinations thereof
[3]:
uberon logical-definitions fingernail toenail
definedClassId: UBERON:0009565
genusIds:
- UBERON:0001705
restrictions:
- fillerId: UBERON:0002389
propertyId: BFO:0000050
---
definedClassId: UBERON:0009567
genusIds:
- UBERON:0001705
restrictions:
- fillerId: UBERON:0001466
propertyId: BFO:0000050
[4]:
uberon logical-definitions fingernail toenail -O csv
[5]:
uberon logical-definitions fingernail toenail -O obo
[Term]
id: UBERON:0009565 ! nail of manual digit
intersection_of: UBERON:0001705 ! nail
intersection_of: BFO:0000050 UBERON:0002389 ! manual digit
[Term]
id: UBERON:0009567 ! nail of pedal digit
intersection_of: UBERON:0001705 ! nail
intersection_of: BFO:0000050 UBERON:0001466 ! pedal digit
Matrix views
We can use the --matrix-axes option to summarize a large collection of logical definitions as a wide table.
This takes two values, separated by a comma:
d: defined_class
f: filler
g: genus
p: predicate
Define class x Predicate
In the following example d,p will create a matrix whose rows are defined classes and whose columns are predicates
[8]:
uberon logical-definitions -p p .desc//p=i "bone element" .and .desc//p=i,p UBERON:0002544 --matrix-axes d,p -O csv -o output/uberon-digit-defs-dp.tsv
[9]:
import pandas as pd
df = pd.read_csv("output/uberon-digit-defs-dp.tsv", sep="\t")
df
[9]:
| defined_class | defined_class_label | genus | genus_label | part_of | part_of_label | |
|---|---|---|---|---|---|---|
| 0 | UBERON:0001436 | phalanx of manus | UBERON:0003221 | phalanx | UBERON:0002389 | manual digit |
| 1 | UBERON:0002234 | proximal phalanx of manus | UBERON:0004302 | proximal phalanx | UBERON:0002389 | manual digit |
| 2 | UBERON:0004328 | proximal phalanx of manual digit 2 | UBERON:0004302 | proximal phalanx | UBERON:0003622 | manual digit 2 |
| 3 | UBERON:0004329 | proximal phalanx of manual digit 3 | UBERON:0004302 | proximal phalanx | UBERON:0003623 | manual digit 3 |
| 4 | UBERON:0004330 | proximal phalanx of manual digit 4 | UBERON:0004302 | proximal phalanx | UBERON:0003624 | manual digit 4 |
| ... | ... | ... | ... | ... | ... | ... |
| 59 | UBERON:0014503 | proximal phalanx of digit 3 | UBERON:0004302 | proximal phalanx | UBERON:0006050 | digit 3 |
| 60 | UBERON:0014504 | proximal phalanx of digit 4 | UBERON:0004302 | proximal phalanx | UBERON:0006051 | digit 4 |
| 61 | UBERON:0014505 | proximal phalanx of digit 5 | UBERON:0004302 | proximal phalanx | UBERON:0006052 | digit 5 |
| 62 | UBERON:0004248 | pedal digit bone | UBERON:0001474 | bone element | UBERON:0001466 | pedal digit |
| 63 | UBERON:0004249 | manual digit bone | UBERON:0001474 | bone element | UBERON:0002389 | manual digit |
64 rows × 6 columns
Filler x Genus
We can flip this around, and have each row be a filler (f) and each column be a genus (g).
[18]:
uberon logical-definitions -p p .desc//p=i "bone element" .and .desc//p=i,p UBERON:0002544 --matrix-axes f,g -O csv -o output/uberon-digit-defs-fg.tsv
[19]:
df = pd.read_csv("output/uberon-digit-defs-fg.tsv", sep="\t")
df
[19]:
| filler | filler_label | phalanx | phalanx_label | proximal_phalanx | proximal_phalanx_label | middle_phalanx | middle_phalanx_label | distal_phalanx | distal_phalanx_label | bone_element | bone_element_label | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | UBERON:0002387 | pes | UBERON:0001449 | phalanx of pes | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 1 | UBERON:0009563 | pastern region of limb | UBERON:0009558 | pastern bone | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 2 | UBERON:0002389 | manual digit | UBERON:0001436 | phalanx of manus | UBERON:0002234 | proximal phalanx of manus | UBERON:0003864 | middle phalanx of manus | UBERON:0003865 | distal phalanx of manus | UBERON:0004249 | manual digit bone |
| 3 | UBERON:0003622 | manual digit 2 | UBERON:0003636 | manual digit 2 phalanx | UBERON:0004328 | proximal phalanx of manual digit 2 | UBERON:0004320 | middle phalanx of manual digit 2 | UBERON:0004311 | distal phalanx of manual digit 2 | NaN | NaN |
| 4 | UBERON:0003623 | manual digit 3 | UBERON:0003637 | manual digit 3 phalanx | UBERON:0004329 | proximal phalanx of manual digit 3 | UBERON:0004321 | middle phalanx of manual digit 3 | UBERON:0004312 | distal phalanx of manual digit 3 | NaN | NaN |
| 5 | UBERON:0003624 | manual digit 4 | UBERON:0003638 | manual digit 4 phalanx | UBERON:0004330 | proximal phalanx of manual digit 4 | UBERON:0004322 | middle phalanx of manual digit 4 | UBERON:0004313 | distal phalanx of manual digit 4 | NaN | NaN |
| 6 | UBERON:0001463 | manual digit 1 | UBERON:0003620 | manual digit 1 phalanx | UBERON:0004338 | proximal phalanx of manual digit 1 | NaN | NaN | UBERON:0004337 | distal phalanx of manual digit 1 | NaN | NaN |
| 7 | UBERON:0003625 | manual digit 5 | UBERON:0003639 | manual digit 5 phalanx | UBERON:0004331 | proximal phalanx of manual digit 5 | UBERON:0004323 | middle phalanx of manual digit 5 | UBERON:0004314 | distal phalanx of manual digit 5 | NaN | NaN |
| 8 | UBERON:0001466 | pedal digit | NaN | NaN | UBERON:0003868 | proximal phalanx of pes | UBERON:0003866 | middle phalanx of pes | UBERON:0003867 | distal phalanx of pes | UBERON:0004248 | pedal digit bone |
| 9 | UBERON:0003632 | pedal digit 2 | UBERON:0003641 | pedal digit 2 phalanx | UBERON:0004333 | proximal phalanx of pedal digit 2 | UBERON:0004324 | middle phalanx of pedal digit 2 | UBERON:0004316 | distal phalanx of pedal digit 2 | NaN | NaN |
| 10 | UBERON:0003633 | pedal digit 3 | UBERON:0003642 | pedal digit 3 phalanx | UBERON:0004334 | proximal phalanx of pedal digit 3 | UBERON:0004325 | middle phalanx of pedal digit 3 | UBERON:0004317 | distal phalanx of pedal digit 3 | NaN | NaN |
| 11 | UBERON:0003634 | pedal digit 4 | UBERON:0003862 | pedal digit 4 phalanx | UBERON:0004335 | proximal phalanx of pedal digit 4 | UBERON:0004326 | middle phalanx of pedal digit 4 | UBERON:0004318 | distal phalanx of pedal digit 4 | NaN | NaN |
| 12 | UBERON:0003631 | pedal digit 1 | UBERON:0003640 | pedal digit 1 phalanx | UBERON:0004332 | proximal phalanx of pedal digit 1 | NaN | NaN | UBERON:0004315 | distal phalanx of pedal digit 1 | NaN | NaN |
| 13 | UBERON:0003635 | pedal digit 5 | UBERON:0003863 | pedal digit 5 phalanx | UBERON:0004336 | proximal phalanx of pedal digit 5 | UBERON:0004327 | middle phalanx of pedal digit 5 | UBERON:0004319 | distal phalanx of pedal digit 5 | NaN | NaN |
| 14 | UBERON:0012137 | pedal digit 7 | UBERON:4100009 | pedal digit 7 phalanx | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 15 | UBERON:0006049 | digit 2 | NaN | NaN | UBERON:0014502 | proximal phalanx of digit 2 | UBERON:0014488 | middle phalanx of digit 2 | UBERON:0014484 | distal phalanx of digit 2 | NaN | NaN |
| 16 | UBERON:0006050 | digit 3 | NaN | NaN | UBERON:0014503 | proximal phalanx of digit 3 | UBERON:0014489 | middle phalanx of digit 3 | UBERON:0014485 | distal phalanx of digit 3 | NaN | NaN |
| 17 | UBERON:0006051 | digit 4 | NaN | NaN | UBERON:0014504 | proximal phalanx of digit 4 | UBERON:0014490 | middle phalanx of digit 4 | UBERON:0014486 | distal phalanx of digit 4 | NaN | NaN |
| 18 | UBERON:0006048 | digit 1 | NaN | NaN | UBERON:0014501 | proximal phalanx of digit 1 | NaN | NaN | UBERON:0014483 | distal phalanx of digit 1 | NaN | NaN |
| 19 | UBERON:0006052 | digit 5 | NaN | NaN | UBERON:0014505 | proximal phalanx of digit 5 | UBERON:0014491 | middle phalanx of digit 5 | UBERON:0014487 | distal phalanx of digit 5 | NaN | NaN |
Note that this view immediately shows the density of the lattice. We can identify what might potentially be gaps;
for example, the cells for “middle phalanx” and digit 1 of the hand (manual) and foot (pedal) are empty. We might think this means we left out a potential term.
However, this omission is actually intentional due to the lack of a middle/intermediate phalanx on the thumb / big toe:
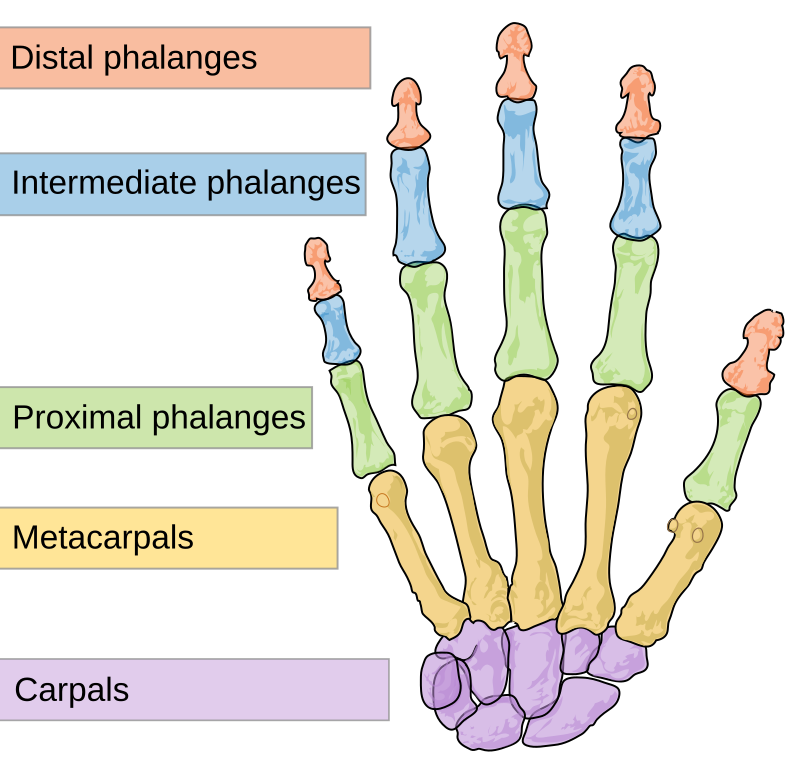
(it may be the case this phalanx is present in other species, in which case a term may be added with negative taxon constraints)
Analyzing and gap filling
OAK has experimental features for analyzing and gap-filling logical definitions; these are not yet exposed on the command line
[ ]: