OAK relationships command
This notebook is intended as a supplement to the main OAK CLI docs.
This notebook provides examples for the relationships command, which can be used to lookup direct and inferred relationships between entities in ontologies.
Overall background on the concepts here can be found in the OAK Guide to Graphs and Relationships.
Help Option
You can get help on any OAK command using --help
[28]:
!runoak relationships --help
Usage: runoak relationships [OPTIONS] [TERMS]...
Show all relationships for a term or terms
By default, this shows all relationships where the input term(s) are the
*subjects*
Example:
runoak -i cl.db relationships CL:4023094
Like all OAK commands, a label can be passed instead of a CURIE
Example:
runoak -i cl.db relationships neuron
To reverse the direction, and query where the search term(s) are *objects*,
use the --direction flag:
Example:
runoak -i cl.db relationships --direction down neuron
Multiple terms can be passed
Example:
runoak -i uberon.db relationships heart liver lung
And like all OAK commands, a query can be passed rather than an explicit
term list
The following query lists all arteries in the limb together which what
structures they supply
Query:
runoak -i uberon.db relationships -p RO:0002178 .desc//p=i "artery" .and
.desc//p=i,p "limb"
More examples:
https://github.com/INCATools/ontology-access-
kit/blob/main/notebooks/Commands/Relationships.ipynb
Python API:
https://incatools.github.io/ontology-access-kit/interfaces/basic
Options:
-p, --predicates TEXT A comma-separated list of predicates. This
may be a shorthand (i, p) or CURIE
--direction [up|down|both] direction of traversal over edges, which up
is subject to object, down is object to
subject.
--autolabel / --no-autolabel If set, results will automatically have
labels assigned [default: autolabel]
-O, --output-type TEXT Desired output type
-o, --output FILENAME Output file, e.g. obo file
--if-absent [absent-only|present-only]
determines behavior when the value is not
present or is empty.
-S, --set-value TEXT the value to set for all terms for the given
property.
--include-entailed / --no-include-entailed
Include entailed indirect relationships
[default: no-include-entailed]
--non-redundant-entailed / --no-non-redundant-entailed
Include entailed but exclude entailed
redundant relationships [default: no-non-
redundant-entailed]
--include-tbox / --no-include-tbox
Include class-class relationships (subclass
and existentials) [default: include-tbox]
--include-abox / --no-include-abox
Include instance relationships (class and
object property assertions) [default:
include-abox]
--include-metadata / --no-include-metadata
Include metadata (axiom annotations)
[default: no-include-metadata]
--help Show this message and exit.
Set up an alias
For convenience we will set up an alias for use in this notebook. This will allow us to use uberon ... rather than runoak -i sqlite:obo:uberon ... for the rest of the notebook.
[2]:
alias uberon runoak -i sqlite:obo:uberon
Direct relationships for a subject term
First we will look up the direct asserted relationships in Uberon with finger as the subject term.
[3]:
uberon relationships finger
subject subject_label predicate predicate_label object object_label
UBERON:0002389 manual digit BFO:0000050 part of UBERON:0002102 forelimb
UBERON:0002389 manual digit BFO:0000050 part of UBERON:0002398 manus
UBERON:0002389 manual digit BFO:0000050 part of UBERON:0012141 manual digitopodium region
UBERON:0002389 manual digit BFO:0000050 part of UBERON:5002389 manual digit plus metapodial segment
UBERON:0002389 manual digit rdfs:subClassOf None UBERON:0002544 digit
Like most OAK commands, the relationships command can take lists of labels, lists of IDs, or even complex query terms (which might themselves involve graphs).
[5]:
uberon relationships finger toe
subject subject_label predicate predicate_label object object_label
UBERON:0001466 pedal digit BFO:0000050 part of UBERON:0002387 pes
UBERON:0001466 pedal digit BFO:0000050 part of UBERON:0012142 pedal digitopodium region
UBERON:0001466 pedal digit BFO:0000050 part of UBERON:5001466 pedal digit plus metapodial segment
UBERON:0001466 pedal digit rdfs:subClassOf None UBERON:0002544 digit
UBERON:0002389 manual digit BFO:0000050 part of UBERON:0002102 forelimb
UBERON:0002389 manual digit BFO:0000050 part of UBERON:0002398 manus
UBERON:0002389 manual digit BFO:0000050 part of UBERON:0012141 manual digitopodium region
UBERON:0002389 manual digit BFO:0000050 part of UBERON:5002389 manual digit plus metapodial segment
UBERON:0002389 manual digit rdfs:subClassOf None UBERON:0002544 digit
Next we will show all direct relationships for all is-a descendants of finger.
[6]:
uberon relationships .desc//p=i finger
subject subject_label predicate predicate_label object object_label
UBERON:0001463 manual digit 1 BFO:0000050 part of UBERON:0002398 manus
UBERON:0001463 manual digit 1 BFO:0000050 part of UBERON:0012141 manual digitopodium region
UBERON:0001463 manual digit 1 BFO:0000050 part of UBERON:5001463 manual digit 1 plus metapodial segment
UBERON:0001463 manual digit 1 BSPO:0001113 preaxialmost part of UBERON:0002398 manus
UBERON:0001463 manual digit 1 rdfs:subClassOf None UBERON:0006048 digit 1
UBERON:0001463 manual digit 1 rdfs:subClassOf None UBERON:0019231 manual digit 1 or 5
UBERON:0002389 manual digit BFO:0000050 part of UBERON:0002102 forelimb
UBERON:0002389 manual digit BFO:0000050 part of UBERON:0002398 manus
UBERON:0002389 manual digit BFO:0000050 part of UBERON:0012141 manual digitopodium region
UBERON:0002389 manual digit BFO:0000050 part of UBERON:5002389 manual digit plus metapodial segment
UBERON:0002389 manual digit rdfs:subClassOf None UBERON:0002544 digit
UBERON:0003622 manual digit 2 BFO:0000050 part of UBERON:0002398 manus
UBERON:0003622 manual digit 2 BFO:0000050 part of UBERON:0012141 manual digitopodium region
UBERON:0003622 manual digit 2 BFO:0000050 part of UBERON:5003622 manual digit 2 plus metapodial segment
UBERON:0003622 manual digit 2 rdfs:subClassOf None UBERON:0006049 digit 2
UBERON:0003622 manual digit 2 rdfs:subClassOf None UBERON:0019232 manual digit 2, 3 or 4
UBERON:0003623 manual digit 3 BFO:0000050 part of UBERON:0002398 manus
UBERON:0003623 manual digit 3 BFO:0000050 part of UBERON:0012141 manual digitopodium region
UBERON:0003623 manual digit 3 BFO:0000050 part of UBERON:5003623 manual digit 3 plus metapodial segment
UBERON:0003623 manual digit 3 rdfs:subClassOf None UBERON:0006050 digit 3
UBERON:0003623 manual digit 3 rdfs:subClassOf None UBERON:0019232 manual digit 2, 3 or 4
UBERON:0003624 manual digit 4 BFO:0000050 part of UBERON:0002398 manus
UBERON:0003624 manual digit 4 BFO:0000050 part of UBERON:0012141 manual digitopodium region
UBERON:0003624 manual digit 4 BFO:0000050 part of UBERON:5003624 manual digit 4 plus metapodial segment
UBERON:0003624 manual digit 4 rdfs:subClassOf None UBERON:0006051 digit 4
UBERON:0003624 manual digit 4 rdfs:subClassOf None UBERON:0019232 manual digit 2, 3 or 4
UBERON:0003625 manual digit 5 BFO:0000050 part of UBERON:0002398 manus
UBERON:0003625 manual digit 5 BFO:0000050 part of UBERON:0012141 manual digitopodium region
UBERON:0003625 manual digit 5 BFO:0000050 part of UBERON:5003625 manual digit 5 plus metapodial segment
UBERON:0003625 manual digit 5 rdfs:subClassOf None UBERON:0006052 digit 5
UBERON:0003625 manual digit 5 rdfs:subClassOf None UBERON:0019231 manual digit 1 or 5
UBERON:0008444 webbed manual digit BFO:0000050 part of UBERON:0008441 webbed manus
UBERON:0008444 webbed manual digit rdfs:subClassOf None UBERON:0002389 manual digit
UBERON:0008444 webbed manual digit rdfs:subClassOf None UBERON:0008443 webbed digit
UBERON:0011981 manual digit 6 BFO:0000050 part of UBERON:5011981 manual digit 6 plus metapodial segment
UBERON:0011981 manual digit 6 rdfs:subClassOf None UBERON:0002389 manual digit
UBERON:0011981 manual digit 6 rdfs:subClassOf None UBERON:0016856 digit 6
UBERON:0011982 manual digit 7 BFO:0000050 part of UBERON:5011982 manual digit 7 plus metapodial segment
UBERON:0011982 manual digit 7 rdfs:subClassOf None UBERON:0002389 manual digit
UBERON:0011982 manual digit 7 rdfs:subClassOf None UBERON:0016857 digit 7
UBERON:0011983 manual digit 8 BFO:0000050 part of UBERON:5011983 manual digit 8 plus metapodial segment
UBERON:0011983 manual digit 8 rdfs:subClassOf None UBERON:0002389 manual digit
UBERON:0011983 manual digit 8 rdfs:subClassOf None UBERON:0016858 digit 8
UBERON:0012260 alular digit BFO:0000050 part of UBERON:5012260 alular digit plus metapodial segment
UBERON:0012260 alular digit BSPO:0001113 preaxialmost part of UBERON:0002398 manus
UBERON:0012260 alular digit RO:0002160 only in taxon NCBITaxon:8782 Aves
UBERON:0012260 alular digit RO:0002254 has developmental contribution from UBERON:0005692 manual digit 2 mesenchyme
UBERON:0012260 alular digit RO:0002254 has developmental contribution from UBERON:0010564 manual digit 1 mesenchyme
UBERON:0012260 alular digit rdfs:subClassOf None UBERON:0002389 manual digit
UBERON:0012261 manual major digit (Aves) BFO:0000050 part of UBERON:5012261 manual major digit (Aves) plus metapodial segment
UBERON:0012261 manual major digit (Aves) RO:0002160 only in taxon NCBITaxon:8782 Aves
UBERON:0012261 manual major digit (Aves) RO:0002254 has developmental contribution from UBERON:0005693 manual digit 3 mesenchyme
UBERON:0012261 manual major digit (Aves) rdfs:subClassOf None UBERON:0002389 manual digit
UBERON:0012262 manual minor digit (Aves) BFO:0000050 part of UBERON:5012262 manual minor digit (Aves) plus metapodial segment
UBERON:0012262 manual minor digit (Aves) BSPO:0001115 postaxialmost part of UBERON:0002398 manus
UBERON:0012262 manual minor digit (Aves) RO:0002160 only in taxon NCBITaxon:8782 Aves
UBERON:0012262 manual minor digit (Aves) RO:0002254 has developmental contribution from UBERON:0005694 manual digit 4 mesenchyme
UBERON:0012262 manual minor digit (Aves) rdfs:subClassOf None UBERON:0002389 manual digit
UBERON:0019231 manual digit 1 or 5 BFO:0000050 part of UBERON:0002398 manus
UBERON:0019231 manual digit 1 or 5 rdfs:subClassOf None UBERON:0002389 manual digit
UBERON:0019231 manual digit 1 or 5 rdfs:subClassOf None UBERON:0019221 digit 1 or 5
UBERON:0019232 manual digit 2, 3 or 4 BFO:0000050 part of UBERON:0002398 manus
UBERON:0019232 manual digit 2, 3 or 4 rdfs:subClassOf None UBERON:0002389 manual digit
UBERON:0019232 manual digit 2, 3 or 4 rdfs:subClassOf None UBERON:0019222 digit 2, 3 or 4
We can write this out to a file and explore it using pandas.
(we use pandas here as this is convenient for Jupyter notebooks but if you were to execute this on the command line you could use any TSV or tabular tool you like)
[7]:
uberon relationships .desc//p=i finger -o output/finger-relationships.tsv
[8]:
import pandas as pd
[9]:
df = pd.read_csv("output/finger-relationships.tsv", sep="\t")
df
[9]:
| subject | subject_label | predicate | predicate_label | object | object_label | |
|---|---|---|---|---|---|---|
| 0 | UBERON:0001463 | manual digit 1 | BFO:0000050 | part of | UBERON:0002398 | manus |
| 1 | UBERON:0001463 | manual digit 1 | BFO:0000050 | part of | UBERON:0012141 | manual digitopodium region |
| 2 | UBERON:0001463 | manual digit 1 | BFO:0000050 | part of | UBERON:5001463 | manual digit 1 plus metapodial segment |
| 3 | UBERON:0001463 | manual digit 1 | BSPO:0001113 | preaxialmost part of | UBERON:0002398 | manus |
| 4 | UBERON:0001463 | manual digit 1 | rdfs:subClassOf | NaN | UBERON:0006048 | digit 1 |
| ... | ... | ... | ... | ... | ... | ... |
| 59 | UBERON:0019231 | manual digit 1 or 5 | rdfs:subClassOf | NaN | UBERON:0002389 | manual digit |
| 60 | UBERON:0019231 | manual digit 1 or 5 | rdfs:subClassOf | NaN | UBERON:0019221 | digit 1 or 5 |
| 61 | UBERON:0019232 | manual digit 2, 3 or 4 | BFO:0000050 | part of | UBERON:0002398 | manus |
| 62 | UBERON:0019232 | manual digit 2, 3 or 4 | rdfs:subClassOf | NaN | UBERON:0002389 | manual digit |
| 63 | UBERON:0019232 | manual digit 2, 3 or 4 | rdfs:subClassOf | NaN | UBERON:0019222 | digit 2, 3 or 4 |
64 rows × 6 columns
Entailments
Next we will look at Entailed relationships.
You are encouraged to consult the OAK guide and glossary here but the basic idea is that entailed relationships encompasses “walking up” the relationship graph, following a specified set of predicates.
First we’ll look at the is-a ancestors of finger. Note the results here should be the same as using the ancestors command:
[10]:
uberon relationships finger --include-entailed -p i
subject subject_label predicate predicate_label object object_label
UBERON:0002389 manual digit rdfs:subClassOf None BFO:0000001 entity
UBERON:0002389 manual digit rdfs:subClassOf None BFO:0000002 continuant
UBERON:0002389 manual digit rdfs:subClassOf None BFO:0000004 independent continuant
UBERON:0002389 manual digit rdfs:subClassOf None BFO:0000040 material entity
UBERON:0002389 manual digit rdfs:subClassOf None UBERON:0000061 anatomical structure
UBERON:0002389 manual digit rdfs:subClassOf None UBERON:0000465 material anatomical entity
UBERON:0002389 manual digit rdfs:subClassOf None UBERON:0000475 organism subdivision
UBERON:0002389 manual digit rdfs:subClassOf None UBERON:0001062 anatomical entity
UBERON:0002389 manual digit rdfs:subClassOf None UBERON:0002389 manual digit
UBERON:0002389 manual digit rdfs:subClassOf None UBERON:0002544 digit
UBERON:0002389 manual digit rdfs:subClassOf None UBERON:0005881 autopodial extension
UBERON:0002389 manual digit rdfs:subClassOf None UBERON:0010000 multicellular anatomical structure
Next we’ll include a wider range of predicates. We’ll also switch our example to be trigeminal ganglion
[18]:
uberon relationships "trigeminal ganglion" --include-entailed -p i,p
subject subject_label predicate predicate_label object object_label
UBERON:0001675 trigeminal ganglion BFO:0000050 part of BFO:0000001 entity
UBERON:0001675 trigeminal ganglion BFO:0000050 part of BFO:0000002 continuant
UBERON:0001675 trigeminal ganglion BFO:0000050 part of BFO:0000004 independent continuant
UBERON:0001675 trigeminal ganglion BFO:0000050 part of BFO:0000040 material entity
UBERON:0001675 trigeminal ganglion BFO:0000050 part of RO:0002577 system
UBERON:0001675 trigeminal ganglion BFO:0000050 part of UBERON:0000033 head
UBERON:0001675 trigeminal ganglion BFO:0000050 part of UBERON:0000061 anatomical structure
UBERON:0001675 trigeminal ganglion BFO:0000050 part of UBERON:0000153 anterior region of body
UBERON:0001675 trigeminal ganglion BFO:0000050 part of UBERON:0000465 material anatomical entity
UBERON:0001675 trigeminal ganglion BFO:0000050 part of UBERON:0000467 anatomical system
UBERON:0001675 trigeminal ganglion BFO:0000050 part of UBERON:0000468 multicellular organism
UBERON:0001675 trigeminal ganglion BFO:0000050 part of UBERON:0000475 organism subdivision
UBERON:0001675 trigeminal ganglion BFO:0000050 part of UBERON:0001016 nervous system
UBERON:0001675 trigeminal ganglion BFO:0000050 part of UBERON:0001062 anatomical entity
UBERON:0001675 trigeminal ganglion BFO:0000050 part of UBERON:0007811 craniocervical region
UBERON:0001675 trigeminal ganglion BFO:0000050 part of UBERON:0010000 multicellular anatomical structure
UBERON:0001675 trigeminal ganglion BFO:0000050 part of UBERON:0011676 subdivision of organism along main body axis
UBERON:0001675 trigeminal ganglion BFO:0000050 part of UBERON:0013701 main body axis
UBERON:0001675 trigeminal ganglion BFO:0000050 part of UBERON:0013702 body proper
UBERON:0001675 trigeminal ganglion rdfs:subClassOf None BFO:0000001 entity
UBERON:0001675 trigeminal ganglion rdfs:subClassOf None BFO:0000002 continuant
UBERON:0001675 trigeminal ganglion rdfs:subClassOf None BFO:0000004 independent continuant
UBERON:0001675 trigeminal ganglion rdfs:subClassOf None BFO:0000040 material entity
UBERON:0001675 trigeminal ganglion rdfs:subClassOf None UBERON:0000045 ganglion
UBERON:0001675 trigeminal ganglion rdfs:subClassOf None UBERON:0000061 anatomical structure
UBERON:0001675 trigeminal ganglion rdfs:subClassOf None UBERON:0000465 material anatomical entity
UBERON:0001675 trigeminal ganglion rdfs:subClassOf None UBERON:0001062 anatomical entity
UBERON:0001675 trigeminal ganglion rdfs:subClassOf None UBERON:0001675 trigeminal ganglion
UBERON:0001675 trigeminal ganglion rdfs:subClassOf None UBERON:0001714 cranial ganglion
UBERON:0001675 trigeminal ganglion rdfs:subClassOf None UBERON:0001800 sensory ganglion
UBERON:0001675 trigeminal ganglion rdfs:subClassOf None UBERON:0004121 ectoderm-derived structure
UBERON:0001675 trigeminal ganglion rdfs:subClassOf None UBERON:0010313 neural crest-derived structure
UBERON:0001675 trigeminal ganglion rdfs:subClassOf None UBERON:0010314 structure with developmental contribution from neural crest
We can see if the query above we get a lot of entailed relationships! Usually we wouldn’t show this as a table to a user - instead we might use the viz command to show all individual direct relationships for all ancestors.
[19]:
uberon viz -p i,p "trigeminal ganglion" -o output/trigeminal-ganglion-graph.png
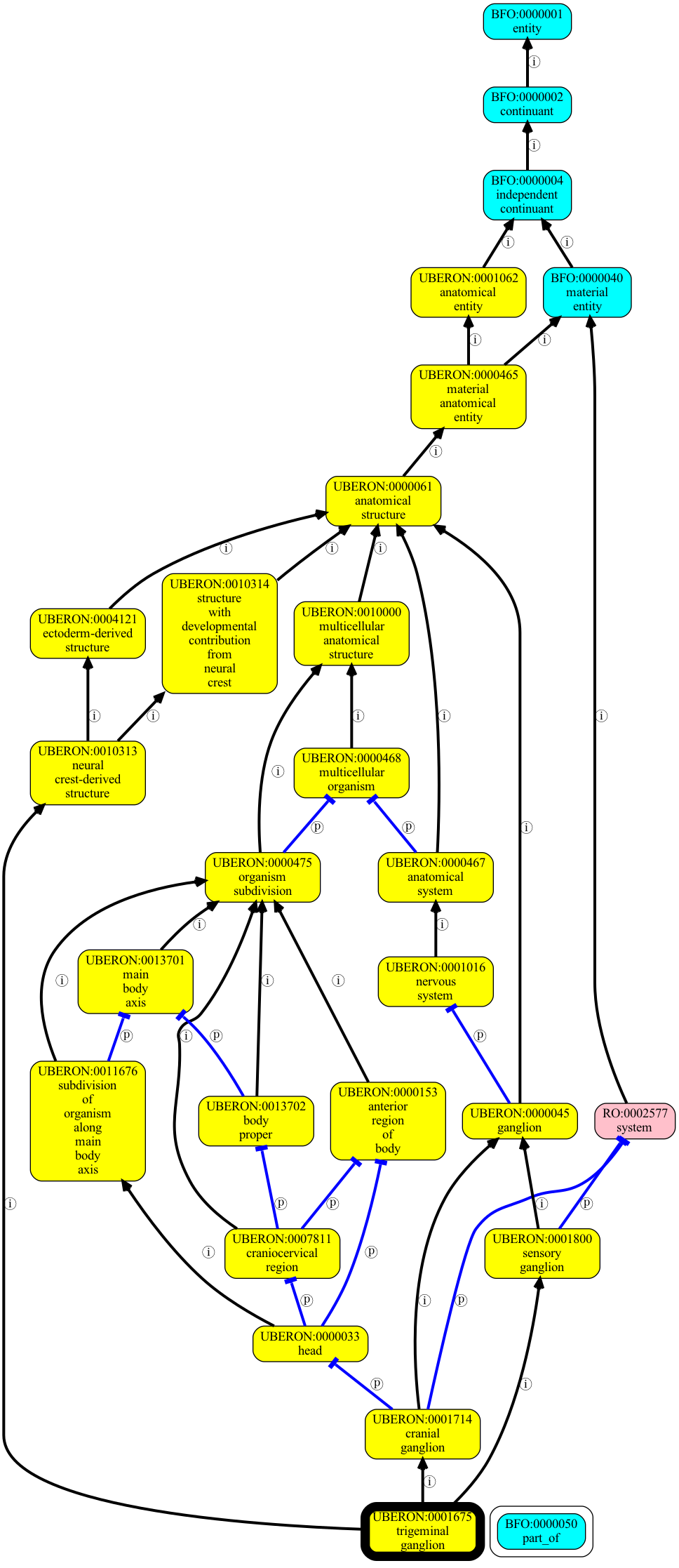
This is a standard way of communicating a complex bundle of relationships. But is there a way of getting the non-redundant informative entailed relationships in a more concise way?
Non-redundant entailed relationships
Is there a way to get the most relevant information in a more concise way, as a table.
Let’s consider the term “trigeminal ganglion” again. Let’s look at direct relationships
[21]:
uberon relationships uberon relationships "trigeminal ganglion" -p i,p
subject subject_label predicate predicate_label object object_label
UBERON:0001675 trigeminal ganglion rdfs:subClassOf None UBERON:0001714 cranial ganglion
UBERON:0001675 trigeminal ganglion rdfs:subClassOf None UBERON:0001800 sensory ganglion
UBERON:0001675 trigeminal ganglion rdfs:subClassOf None UBERON:0010313 neural crest-derived structure
These are all correct but don’t tell us what this ganglion is a part of. Using the --include-entailed option above gives too much information.
OAK now has a --non-redundant-entailed option which effectively “rolls down” the entailed relationships for each predicate:
[22]:
uberon relationships uberon relationships --non-redundant-entailed "trigeminal ganglion" -p i,p
subject subject_label predicate predicate_label object object_label
UBERON:0001675 trigeminal ganglion BFO:0000050 part of RO:0002577 system
UBERON:0001675 trigeminal ganglion BFO:0000050 part of UBERON:0000033 head
UBERON:0001675 trigeminal ganglion BFO:0000050 part of UBERON:0001016 nervous system
UBERON:0001675 trigeminal ganglion rdfs:subClassOf None UBERON:0001714 cranial ganglion
UBERON:0001675 trigeminal ganglion rdfs:subClassOf None UBERON:0001800 sensory ganglion
UBERON:0001675 trigeminal ganglion rdfs:subClassOf None UBERON:0010313 neural crest-derived structure
Note that even though 3 part-parents are provided, these are all technically non-redundant, as they are all “proper” overlaps (the system term is odd, but this is an artefact of RO imports, and in fact uberon doesn’t place ‘nervous system’ under ‘system’)
We can do this for other relationships too:
[26]:
uberon relationships uberon relationships --non-redundant-entailed "trigeminal ganglion"
subject subject_label predicate predicate_label object object_label
UBERON:0001675 trigeminal ganglion BFO:0000050 part of RO:0002577 system
UBERON:0001675 trigeminal ganglion BFO:0000051 has part UBERON:0003714 neural tissue
UBERON:0001675 trigeminal ganglion RO:0002131 overlaps RO:0002577 system
UBERON:0001675 trigeminal ganglion RO:0002131 overlaps UBERON:0003714 neural tissue
UBERON:0001675 trigeminal ganglion RO:0002162 in taxon NCBITaxon:7742 Vertebrata <vertebrates>
UBERON:0001675 trigeminal ganglion RO:0002170 connected to UBERON:0001027 sensory nerve
UBERON:0001675 trigeminal ganglion RO:0002170 connected to UBERON:0001645 trigeminal nerve
UBERON:0001675 trigeminal ganglion RO:0002202 develops from UBERON:0006304 future trigeminal ganglion
UBERON:0001675 trigeminal ganglion RO:0002207 directly develops from UBERON:0006304 future trigeminal ganglion
UBERON:0001675 trigeminal ganglion RO:0002225 develops from part of UBERON:0000922 embryo
UBERON:0001675 trigeminal ganglion RO:0002254 has developmental contribution from UBERON:0006304 future trigeminal ganglion
UBERON:0001675 trigeminal ganglion RO:0002258 developmentally preceded by UBERON:0006304 future trigeminal ganglion
UBERON:0001675 trigeminal ganglion RO:0002328 functionally related to GO:0019226 transmission of nerve impulse
UBERON:0001675 trigeminal ganglion RO:0002329 part of structure that is capable of GO:0050877 nervous system process
UBERON:0001675 trigeminal ganglion RO:0002473 composed primarily of UBERON:0003714 neural tissue
UBERON:0001675 trigeminal ganglion RO:0002494 transformation of UBERON:0006304 future trigeminal ganglion
UBERON:0001675 trigeminal ganglion RO:0002495 immediate transformation of UBERON:0006304 future trigeminal ganglion
UBERON:0001675 trigeminal ganglion RO:0002496 existence starts during or after UBERON:0000110 neurula stage
UBERON:0001675 trigeminal ganglion RO:0002496 existence starts during or after UBERON:0000111 organogenesis stage
UBERON:0001675 trigeminal ganglion RO:0002497 existence ends during or before UBERON:0000066 fully formed stage
UBERON:0001675 trigeminal ganglion RO:0002584 has part structure that is capable of GO:0019226 transmission of nerve impulse
UBERON:0001675 trigeminal ganglion rdfs:subClassOf None UBERON:0001714 cranial ganglion
UBERON:0001675 trigeminal ganglion rdfs:subClassOf None UBERON:0001800 sensory ganglion
UBERON:0001675 trigeminal ganglion rdfs:subClassOf None UBERON:0010313 neural crest-derived structure
UBERON:0001675 trigeminal ganglion uberon/core#extends:fibers_into None UBERON:0001027 sensory nerve
UBERON:0001675 trigeminal ganglion uberon/core#extends:fibers_into None UBERON:0001645 trigeminal nerve
[ ]: