command: crawl
This notebook is intended as a supplement to the main OAK CLI docs.
This notebook provides examples for the crawl command, which is used to walk over multiple ontologies and endpoints.
Help Option
You can get help on any OAK command using --help
[1]:
!runoak crawl --help
Usage: runoak crawl [OPTIONS] [TERMS]...
Crawl one or more ontologies, hopping over edges and mappings
Crawl is a powerful command that allows for multi-ontology traversal,
particularly on mapping paths. Multiple ontologies and ontology sources
(e.g. BioPortal, OLS) provide mappings between terms. No single ontology is
likely to have a complete source. Using crawl, you can walk across the union
of mappings in all ontologies, with custom rules for each ontology (e.g.
normalizing prefixes).
Documentation for this command will be provided in a separate notebook.
Options:
-o, --output TEXT Path to output file
-O, --output-type TEXT Desired output type
--autolabel / --no-autolabel If set, results will automatically have
labels assigned [default: autolabel]
-M, --maps-to-source TEXT Return only mappings with subject or object
source equal to this
--mapper TEXT A selector for an adapter that is to be used
for the main lookup operation
--unmelt / --no-unmelt Use a wide table for display. [default: no-
unmelt]
--adapters TEXT A comma-separated list of adapters
--allowed-prefixes TEXT A comma-separated list of prefixes to
traverse over
--mapping-predicates TEXT A comma-separated list of mapping predicates
to traverse over
--viz / --no-viz If true then draw a graph [default: no-viz]
-d, --directory TEXT Directory to write output files
--whole-ontology / --no-whole-ontology
Run over whole ontology [default: no-whole-
ontology]
-C, --config-yaml TEXT
--help Show this message and exit.
[2]:
!cat input/mapping-crawler-config.yaml
adapter_configs:
OMIM:
DOID:
prefix_normalization_map:
"MIM:PS": "OMIMPS:"
MIM: OMIM
UMLS_CUI: UMLS
SNOMEDCT_US_2023_03_01: SCTID
NCI: NCIT
ORDO:
prefix_normalization_map:
MeSH: MESH
NCIT:
GARD:
adapter_specs:
OMIM: /Users/cjm/repos/semantic-sql/db/omim.db
allowed_prefixes: [OMIM, DOID, ORDO, OMIMPS, GARD, NCIT, OMIMPS]
mapping_predicates:
- oio:hasDbXref
- skos:exactMatch
[4]:
!runoak crawl -C input/mapping-crawler-config.yaml -d output/refsum-analysis GARD:4648 --viz -o output/refsum.png
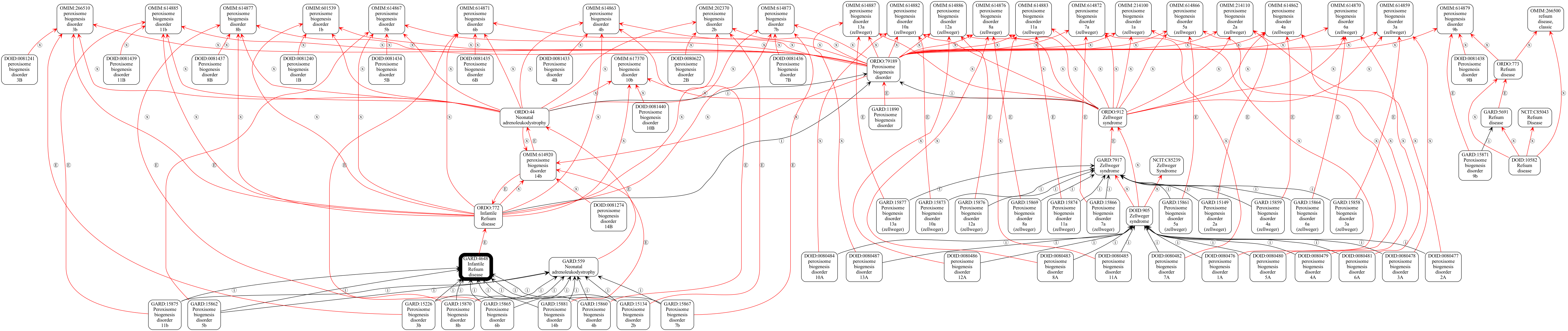
[8]:
!runoak crawl -C input/mapping-crawler-config.yaml -d output/refsum-analysis --whole-ontology GARD:4648 GARD:6322
/Users/cjm/Library/Caches/pypoetry/virtualenvs/oaklib-OeQZizwE-py3.9/lib/python3.9/site-packages/sssom/util.py:162: FutureWarning: Downcasting behavior in `replace` is deprecated and will be removed in a future version. To retain the old behavior, explicitly call `result.infer_objects(copy=False)`. To opt-in to the future behavior, set `pd.set_option('future.no_silent_downcasting', True)`
df.replace("", np.nan, inplace=True)
/Users/cjm/Library/Caches/pypoetry/virtualenvs/oaklib-OeQZizwE-py3.9/lib/python3.9/site-packages/sssom/util.py:162: FutureWarning: Downcasting behavior in `replace` is deprecated and will be removed in a future version. To retain the old behavior, explicitly call `result.infer_objects(copy=False)`. To opt-in to the future behavior, set `pd.set_option('future.no_silent_downcasting', True)`
df.replace("", np.nan, inplace=True)
[9]:
!find output/refsum-analysis
output/refsum-analysis
output/refsum-analysis/clique_summary.csv
output/refsum-analysis/clique_results.csv
output/refsum-analysis/cliques
output/refsum-analysis/cliques/GARD_4648.sssom.tsv
output/refsum-analysis/cliques/GARD_6322.sssom.tsv
[10]:
import pandas as pd
[12]:
pd.read_csv("output/refsum-analysis/clique_summary.csv")
[12]:
| Unnamed: 0 | mapping_count | entity_count | average_incoherency | max_incoherency | incoherency_GARD | incoherency_ORDO | incoherency_OMIM | incoherency_DOID | incoherency_NCIT | incoherency_OMIMPS | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | count | 2.000000 | 2.000000 | 2.000000 | 2.00000 | 2.00000 | 2.000000 | 1.0 | 2.00000 | 2.000000 | 1.0 |
| 1 | mean | 145.500000 | 44.500000 | 7.900000 | 12.50000 | 12.50000 | 2.000000 | 24.0 | 12.50000 | 0.500000 | 0.0 |
| 2 | std | 194.454365 | 55.861436 | 11.172287 | 17.67767 | 17.67767 | 2.828427 | NaN | 17.67767 | 0.707107 | NaN |
| 3 | min | 8.000000 | 5.000000 | 0.000000 | 0.00000 | 0.00000 | 0.000000 | 24.0 | 0.00000 | 0.000000 | 0.0 |
| 4 | 25% | 76.750000 | 24.750000 | 3.950000 | 6.25000 | 6.25000 | 1.000000 | 24.0 | 6.25000 | 0.250000 | 0.0 |
| 5 | 50% | 145.500000 | 44.500000 | 7.900000 | 12.50000 | 12.50000 | 2.000000 | 24.0 | 12.50000 | 0.500000 | 0.0 |
| 6 | 75% | 214.250000 | 64.250000 | 11.850000 | 18.75000 | 18.75000 | 3.000000 | 24.0 | 18.75000 | 0.750000 | 0.0 |
| 7 | max | 283.000000 | 84.000000 | 15.800000 | 25.00000 | 25.00000 | 4.000000 | 24.0 | 25.00000 | 1.000000 | 0.0 |
[13]:
pd.read_csv("output/refsum-analysis/clique_results.csv")
[13]:
| seed | name | mapping_count | entity_count | entities | entity_labels | predicates | mapping_sources | average_incoherency | max_incoherency | sources | incoherency_GARD | incoherency_ORDO | incoherency_OMIM | incoherency_DOID | incoherency_NCIT | incoherency_OMIMPS | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | GARD:4648 | NaN | 283 | 84 | ['OMIM:614867', 'ORDO:772', 'OMIM:614873', 'GA... | {'GARD:4648': 'Infantile Refsum disease', 'ORD... | ['rdfs:subClassOf', 'oio:hasDbXref', 'skos:exa... | ['obo:ORDO', 'obo:GARD', 'obo:DOID', 'obo:OMIM'] | 15.8 | 25 | ['GARD', 'ORDO', 'OMIM', 'DOID', 'NCIT'] | 25 | 4 | 24.0 | 25 | 1 | NaN |
| 1 | GARD:6322 | NaN | 8 | 5 | ['ORDO:98249', 'DOID:13359', 'NCIT:C34568', 'O... | {'DOID:13359': 'Ehlers-Danlos syndrome', 'GARD... | ['oio:hasDbXref', 'skos:exactMatch'] | ['obo:DOID', 'obo:GARD'] | 0.0 | 0 | ['DOID', 'GARD', 'ORDO', 'OMIMPS', 'NCIT'] | 0 | 0 | NaN | 0 | 0 | 0.0 |
[15]:
pd.read_csv("output/refsum-analysis/cliques/GARD_4648.sssom.tsv", sep="\t", comment="#")
[15]:
| subject_id | subject_label | predicate_id | object_id | object_label | mapping_justification | mapping_source | other | |
|---|---|---|---|---|---|---|---|---|
| 0 | DOID:0080476 | peroxisome biogenesis disorder 1A | oio:hasDbXref | OMIM:214100 | peroxisome biogenesis disorder 1a (zellweger) | semapv:UnspecifiedMatching | obo:DOID | distance: 6, direction: -1 |
| 1 | DOID:0080476 | peroxisome biogenesis disorder 1A | oio:hasDbXref | OMIM:214100 | peroxisome biogenesis disorder 1a (zellweger) | semapv:UnspecifiedMatching | obo:DOID | distance: 7, direction: 1 |
| 2 | DOID:0080476 | peroxisome biogenesis disorder 1A | rdfs:subClassOf | DOID:905 | Zellweger syndrome | semapv:ManualMappingCuration | obo:DOID | NaN |
| 3 | DOID:0080477 | peroxisome biogenesis disorder 2A | oio:hasDbXref | OMIM:214110 | peroxisome biogenesis disorder 2a (zellweger) | semapv:UnspecifiedMatching | obo:DOID | distance: 6, direction: -1 |
| 4 | DOID:0080477 | peroxisome biogenesis disorder 2A | oio:hasDbXref | OMIM:214110 | peroxisome biogenesis disorder 2a (zellweger) | semapv:UnspecifiedMatching | obo:DOID | distance: 7, direction: 1 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 278 | ORDO:912 | Zellweger syndrome | oio:hasDbXref | OMIM:614887 | peroxisome biogenesis disorder 13a (zellweger) | semapv:UnspecifiedMatching | obo:ORDO | distance: 3, direction: 1 |
| 279 | ORDO:912 | Zellweger syndrome | oio:hasDbXref | OMIM:614887 | peroxisome biogenesis disorder 13a (zellweger) | semapv:UnspecifiedMatching | obo:ORDO | distance: 4, direction: -1 |
| 280 | ORDO:912 | Zellweger syndrome | oio:hasDbXref | OMIM:617370 | peroxisome biogenesis disorder 10b | semapv:UnspecifiedMatching | obo:ORDO | distance: 2, direction: -1 |
| 281 | ORDO:912 | Zellweger syndrome | oio:hasDbXref | OMIM:617370 | peroxisome biogenesis disorder 10b | semapv:UnspecifiedMatching | obo:ORDO | distance: 3, direction: 1 |
| 282 | ORDO:912 | Zellweger syndrome | rdfs:subClassOf | ORDO:79189 | Peroxisome biogenesis disorder | semapv:ManualMappingCuration | obo:ORDO | NaN |
283 rows × 8 columns
[ ]: