OAK diff-via-mappings command
This notebook is intended as a supplement to the main OAK CLI docs.
This notebook provides examples for the diff-via-mappings command, which can be used to find differences between two ontologies based on mappings between them
For more on mappings, see Mappings in the OAK guide.
Help Option
You can get help on any OAK command using --help
[1]:
!runoak diff-via-mappings --help
Usage: runoak diff-via-mappings [OPTIONS] [TERMS]...
Calculates cross-ontology diff using mappings
Given a pair of ontologies, and mappings that connect terms in both
ontologies, this command will perform a structural comparison of all mapped
pairs of terms
Example:
runoak -i sqlite:obo:uberon diff-via-mappings --other-input
sqlite:obo:zfa --source UBERON --source ZFA -O csv
Note the above command does not have any mapping file specified; the
mappings that are distributed within each ontology is used (in this case,
Uberon contains mappings to ZFA)
If the mappings are provided externally:
runoak -i ont1.obo diff-via-mappings --other-input ont2.obo --mapping-
input mappings.sssom.tsv
(in the above example, --source is not passed, so all mappings are tested)
If there are no existing mappings, you can use the lexmatch command to
generate them:
runoak -i ont1.obo diff-via-mappings -a ont2.obo lexmatch -o
mappings.sssom.tsv
runoak -i ont1.obo diff-via-mappings --other-input ont2.obo --mapping-
input mappings.sssom.tsv
The output from this command follows the cross-ontology-diff data model
(https://incatools.github.io/ontology-access-kit/datamodels/cross-ontology-
diff/index.html)
This can be serialized in YAML or TSV form
Options:
-S, --source TEXT ontology prefixes e.g. HP, MP
--mapping-input TEXT File of mappings in SSSOM format. If not
provided then mappings in ontology(ies) are
used
-X, --other-input TEXT Additional input file
--other-input-type TEXT Type of additional input file
--intra / --no-intra If true, then all sources are in the main
input ontology [default: no-intra]
--autolabel / --no-autolabel If set, results will automatically have
labels assigned [default: autolabel]
--include-identity-mappings / --no-include-identity-mappings
Use identity relation as mapping; use this
for two versions of the same ontology
[default: no-include-identity-mappings]
--filter-category-identical / --no-filter-category-identical
Do not report cases where a relationship has
not changed [default: no-filter-category-
identical]
--bidirectional / --no-bidirectional
Show diff from both left and right
perspectives [default: bidirectional]
-p, --predicates TEXT A comma-separated list of predicates. This
may be a shorthand (i, p) or CURIE
-o, --output FILENAME Output file, e.g. obo file
-O, --output-type TEXT Desired output type
--help Show this message and exit.
Example: Diff between two anatomy ontologies
To illustrate usage, we will calculate the diff between UBERON (a multi-species anatomy ontology) and ZFA (an anatomy ontology for zebrafish).
Note that rather than provide a set of external mappings, we will use the mappings present in both ontologies (in this case, UBERON has xrefs to ZFA).
To simplify the comparison, we will only consider is_a and part_of relationships. This is specified using the --predicates (-p) option
[5]:
!runoak -i sqlite:obo:uberon diff-via-mappings --other-input \
sqlite:obo:zfa --source UBERON --source ZFA -p i,p -O csv -o output/uberon-zfa-diff.csv
Analyzing the results using Pandas
Note that we asked for the output as a tabular file (-O csv), rather than native YAML. You could take the tabular output, analyze it in a spreadsheet, etc. Here we will use the Python pandas library.
[8]:
import pandas as pd
df=pd.read_csv("output/uberon-zfa-diff.csv", sep="\t").fillna("")
[9]:
df
[9]:
| left_subject_id | left_object_id | left_predicate_id | category | left_subject_label | left_object_label | left_predicate_label | right_subject_id | right_object_id | right_predicate_ids | right_subject_label | right_object_label | right_predicate_labels | left_subject_is_functional | left_object_is_functional | subject_mapping_predicate | object_mapping_predicate | right_intermediate_ids | subject_mapping_cardinality | object_mapping_cardinality | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | UBERON:0000002 | UBERON:0000995 | BFO:0000050 | MissingMapping | uterine cervix | uterus | 1:0 | |||||||||||||
| 1 | UBERON:0000002 | UBERON:0001560 | rdfs:subClassOf | MissingMapping | uterine cervix | neck of organ | 1:0 | |||||||||||||
| 2 | UBERON:0000002 | UBERON:0005156 | rdfs:subClassOf | MissingMapping | uterine cervix | reproductive structure | 1:0 | |||||||||||||
| 3 | UBERON:0000003 | UBERON:0000033 | BFO:0000050 | MissingMapping | naris | head | 1:0 | |||||||||||||
| 4 | UBERON:0000003 | UBERON:0005725 | BFO:0000050 | MissingMapping | naris | olfactory system | 1:0 | |||||||||||||
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 36336 | ZFA:0009401 | ZFA:0009000 | rdfs:subClassOf | MissingMapping | lens fiber cell | cell | 1:0 | |||||||||||||
| 36337 | ZFA:0009402 | ZFA:0005065 | BFO:0000050 | MissingMapping | heart valve cell | heart valve | 1:0 | |||||||||||||
| 36338 | ZFA:0009402 | ZFA:0009000 | rdfs:subClassOf | MissingMapping | heart valve cell | cell | 1:0 | |||||||||||||
| 36339 | ZFA:0009403 | ZFA:0009402 | rdfs:subClassOf | MissingMapping | heart valve interstitial cell | heart valve cell | 1:0 | |||||||||||||
| 36340 | ZFA:0009404 | ZFA:0009402 | rdfs:subClassOf | MissingMapping | heart valve endothelial cell | heart valve cell | 1:0 |
36341 rows × 20 columns
[20]:
df["left_source"] = df.apply(lambda x: x.left_subject_id.split(":")[0], axis=1)
Plotting mapping diff categories
The diff tool works by taking each relationship/edge in the “left” ontology (here all UBERON and all ZFA edges are considered) and trying to map it to a relationship in the “right” ontology.
The mapping is assigned a category from the cross-ontology-diff:DiffCategory enumeration.
(note that like many OAK operations, the output conforms to a data model that makes its semantics explicit)
We will use seaborn/matplotlib to plot the category counts
[11]:
import seaborn as sns
import matplotlib.pyplot as plt
# set plot style: grey grid in the background:
sns.set(style="darkgrid")
# Set the figure size
plt.figure(figsize=(10, 7))
sns.countplot(data=df, x='category')
plt.title('Count of each category')
plt.xlabel('Category')
plt.ylabel('Count')
plt.show()
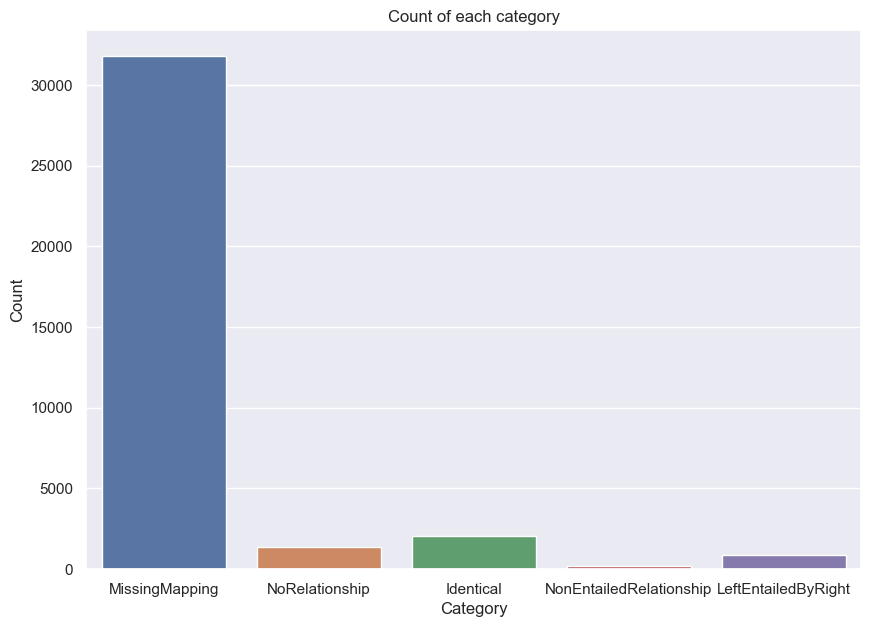
the dominant category is MissingMapping. This means that either/both subject and object of the left edge could not be matched in the right ontology. It is not surprising there are so many, because Uberon covers more species.
Breaking down by predicate, and using log scale
We can get a more detailed view by breaking down the counts by the predicate of the left edge.
We will also use a log scale
[14]:
sns.countplot(data=df, x='category', hue="left_predicate_id")
plt.title('Count of each category')
plt.xlabel('Category')
plt.ylabel('Count')
plt.yscale('log')
plt.xticks(rotation=45) # Make x-axis labels diagonal
plt.show()
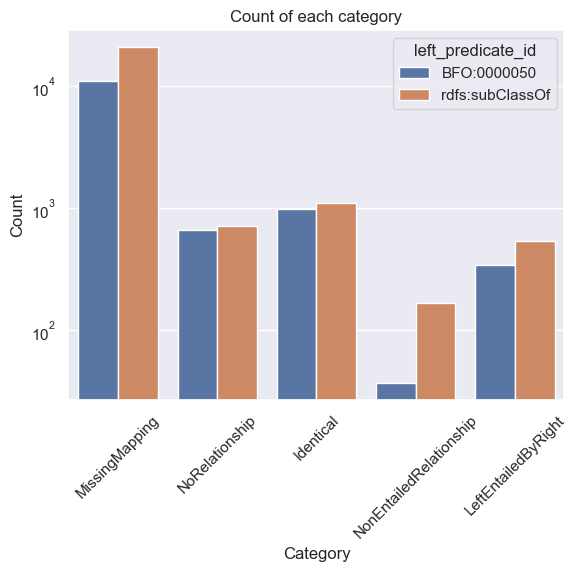
Exploring categories
Non-entailed relationships
i.e. this is an edge in UBERON that corresponds to a different kind of edge in ZFA (or vice versa), where the former doesn’t subsume the latter.
For example, in UBERON PNS part_of NS; in ZFA PNS is_a NS
[17]:
df.query("category=='NonEntailedRelationship'")
[17]:
| left_subject_id | left_object_id | left_predicate_id | category | left_subject_label | left_object_label | left_predicate_label | right_subject_id | right_object_id | right_predicate_ids | right_subject_label | right_object_label | right_predicate_labels | left_subject_is_functional | left_object_is_functional | subject_mapping_predicate | object_mapping_predicate | right_intermediate_ids | subject_mapping_cardinality | object_mapping_cardinality | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 22 | UBERON:0000010 | UBERON:0001016 | BFO:0000050 | NonEntailedRelationship | peripheral nervous system | nervous system | ZFA:0000142 | ZFA:0000396 | rdfs:subClassOf | peripheral nervous system | nervous system | |||||||||
| 24 | UBERON:0000011 | UBERON:0002410 | BFO:0000050 | NonEntailedRelationship | parasympathetic nervous system | autonomic nervous system | ZFA:0001575 | ZFA:0001574 | rdfs:subClassOf | parasympathetic nervous system | autonomic nervous system | |||||||||
| 29 | UBERON:0000013 | UBERON:0002410 | BFO:0000050 | NonEntailedRelationship | sympathetic nervous system | autonomic nervous system | ZFA:0001576 | ZFA:0001574 | rdfs:subClassOf | sympathetic nervous system | autonomic nervous system | |||||||||
| 196 | UBERON:0000095 | UBERON:0002342 | BFO:0000050 | NonEntailedRelationship | cardiac neural crest | neural crest | ZFA:0001648 | ZFA:0000045 | rdfs:subClassOf | cardiac neural crest | neural crest | |||||||||
| 725 | UBERON:0000936 | UBERON:0003931 | BFO:0000050 | NonEntailedRelationship | posterior commissure | diencephalic white matter | ZFA:0000320 | ZFA:0000338 | rdfs:subClassOf | caudal commissure | diencephalic white matter | |||||||||
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 35155 | ZFA:0005661 | ZFA:0001488 | rdfs:subClassOf | NonEntailedRelationship | olfactory bulb glomerulus | multi-tissue structure | UBERON:0005387 | UBERON:0000481 | BFO:0000050|RO:0002131|RO:0002323 | olfactory glomerulus | multi-tissue structure | |||||||||
| 35458 | ZFA:0005829 | ZFA:0005249 | rdfs:subClassOf | NonEntailedRelationship | Schlemm's canal | vasculature | UBERON:0004029 | UBERON:0002049 | BFO:0000050|RO:0002131|RO:0002323 | canal of Schlemm | vasculature | |||||||||
| 35680 | ZFA:0007017 | ZFA:0001477 | rdfs:subClassOf | NonEntailedRelationship | posterior neural plate | portion of tissue | UBERON:0003057 | UBERON:0000479 | BFO:0000050|RO:0002131|RO:0002323 | chordal neural plate | tissue | |||||||||
| 35701 | ZFA:0007037 | ZFA:0001477 | rdfs:subClassOf | NonEntailedRelationship | posterior neural tube | portion of tissue | UBERON:0003076 | UBERON:0000479 | BFO:0000050|RO:0002131|RO:0002202|RO:0002254|R... | posterior neural tube | tissue | |||||||||
| 35702 | ZFA:0007038 | ZFA:0001477 | rdfs:subClassOf | NonEntailedRelationship | anterior neural tube | portion of tissue | UBERON:0003080 | UBERON:0000479 | BFO:0000050|RO:0002131|RO:0002202|RO:0002254|R... | anterior neural tube | tissue |
205 rows × 20 columns
No relationship
In this case, the edge in UBERON has no corresponding edge in ZFA (or vice versa), entailed or otherwise
[18]:
df.query("category=='NoRelationship'")
[18]:
| left_subject_id | left_object_id | left_predicate_id | category | left_subject_label | left_object_label | left_predicate_label | right_subject_id | right_object_id | right_predicate_ids | right_subject_label | right_object_label | right_predicate_labels | left_subject_is_functional | left_object_is_functional | subject_mapping_predicate | object_mapping_predicate | right_intermediate_ids | subject_mapping_cardinality | object_mapping_cardinality | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 7 | UBERON:0000004 | UBERON:0000475 | rdfs:subClassOf | NoRelationship | nose | organism subdivision | ZFA:0000047 | ZFA:0001308 | peripheral olfactory organ | organism subdivision | ||||||||||
| 188 | UBERON:0000089 | UBERON:0004750 | BFO:0000050 | NoRelationship | hypoblast (generic) | blastoderm | ZFA:0000117 | ZFA:0001176 | hypoblast | blastoderm | ||||||||||
| 647 | UBERON:0000471 | UBERON:0003103 | BFO:0000050 | NoRelationship | compound organ component | compound organ | ZFA:0001489 | ZFA:0000496 | compound organ component | compound organ | ||||||||||
| 661 | UBERON:0000479 | UBERON:0000468 | BFO:0000050 | NoRelationship | tissue | multicellular organism | ZFA:0001477 | ZFA:0001094 | portion of tissue | whole organism | ||||||||||
| 663 | UBERON:0000480 | UBERON:0000468 | BFO:0000050 | NoRelationship | anatomical group | multicellular organism | ZFA:0001512 | ZFA:0001094 | anatomical group | whole organism | ||||||||||
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 35714 | ZFA:0007057 | ZFA:0005297 | rdfs:subClassOf | NoRelationship | ocular blood vessel | cranial blood vessel | UBERON:0002203 | UBERON:0011362 | vasculature of eye | cranial blood vasculature | ||||||||||
| 35715 | ZFA:0007058 | ZFA:0000012 | BFO:0000050 | NoRelationship | roof plate | central nervous system | UBERON:0003054 | UBERON:0001017 | roof plate | central nervous system | ||||||||||
| 35716 | ZFA:0007058 | ZFA:0001488 | rdfs:subClassOf | NoRelationship | roof plate | multi-tissue structure | UBERON:0003054 | UBERON:0000481 | roof plate | multi-tissue structure | ||||||||||
| 35720 | ZFA:0007071 | ZFA:0001477 | rdfs:subClassOf | NoRelationship | flexural organ | portion of tissue | UBERON:0011577 | UBERON:0000479 | flexural organ | tissue | ||||||||||
| 35722 | ZFA:0007072 | ZFA:0001488 | rdfs:subClassOf | NoRelationship | blood sinus | multi-tissue structure | UBERON:0006615 | UBERON:0000481 | venous sinus | multi-tissue structure |
1375 rows × 20 columns
LeftEntailedByRight
in this case UBERON has a direct edge that corresponds to two or more direct edges in ZFA (or vice versa)
In this case the edges must chain together via OWL semantics.
for example, UBERON has ganglion part-of nervous system, ZFA has ganglion part-of PNS is-a nervous system
[25]:
df.query("category=='LeftEntailedByRight'")
[25]:
| left_subject_id | left_object_id | left_predicate_id | category | left_subject_label | left_object_label | left_predicate_label | right_subject_id | right_object_id | right_predicate_ids | ... | right_object_label | right_predicate_labels | left_subject_is_functional | left_object_is_functional | subject_mapping_predicate | object_mapping_predicate | right_intermediate_ids | subject_mapping_cardinality | object_mapping_cardinality | left_source | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 35 | UBERON:0000016 | UBERON:0000949 | BFO:0000050 | LeftEntailedByRight | endocrine pancreas | endocrine system | ZFA:0001260 | ZFA:0001158 | BFO:0000050|RO:0002131|RO:0002202 | ... | endocrine system | UBERON | |||||||||
| 91 | UBERON:0000045 | UBERON:0001016 | BFO:0000050 | LeftEntailedByRight | ganglion | nervous system | ZFA:0000190 | ZFA:0000396 | BFO:0000050|RO:0002131 | ... | nervous system | UBERON | |||||||||
| 182 | UBERON:0000086 | UBERON:0000992 | BFO:0000050 | LeftEntailedByRight | zona pellucida | ovary | ZFA:0001111 | ZFA:0000403 | BFO:0000050|RO:0002131 | ... | ovary | UBERON | |||||||||
| 645 | UBERON:0000467 | UBERON:0000061 | rdfs:subClassOf | LeftEntailedByRight | anatomical system | anatomical structure | ZFA:0001439 | ZFA:0000037 | BFO:0000050|RO:0002131|rdfs:subClassOf | ... | anatomical structure | UBERON | |||||||||
| 658 | UBERON:0000477 | UBERON:0001062 | rdfs:subClassOf | LeftEntailedByRight | anatomical cluster | anatomical entity | ZFA:0001478 | ZFA:0100000 | rdfs:subClassOf | ... | zebrafish anatomical entity | UBERON | |||||||||
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 35706 | ZFA:0007043 | ZFA:0001477 | rdfs:subClassOf | LeftEntailedByRight | hindbrain neural tube | portion of tissue | UBERON:2007043 | UBERON:0000479 | BFO:0000050|RO:0002131|RO:0002202|RO:0002254|R... | ... | tissue | ZFA | |||||||||
| 35711 | ZFA:0007048 | ZFA:0005145 | rdfs:subClassOf | LeftEntailedByRight | ventral intermandibularis posterior | muscle | UBERON:2007048 | UBERON:0001630 | RO:0002131|RO:0002202|RO:0002254|RO:0002258|RO... | ... | muscle organ | ZFA | |||||||||
| 35717 | ZFA:0007068 | ZFA:0001486 | rdfs:subClassOf | LeftEntailedByRight | otic epithelium | epithelium | UBERON:0003249 | UBERON:0000483 | RO:0002131|RO:0002323|rdfs:subClassOf | ... | epithelium | ZFA | |||||||||
| 35721 | ZFA:0007072 | ZFA:0000010 | BFO:0000050 | LeftEntailedByRight | blood sinus | cardiovascular system | UBERON:0006615 | UBERON:0004535 | BFO:0000050|RO:0002131|RO:0002202|RO:0002254|R... | ... | cardiovascular system | ZFA | |||||||||
| 35723 | ZFA:0007073 | ZFA:0001643 | rdfs:subClassOf | LeftEntailedByRight | blood sinus cavity | anatomical space | UBERON:0034940 | UBERON:0000464 | rdfs:subClassOf | ... | anatomical space | ZFA |
883 rows × 21 columns
Breaking things down by direction
By default, the command will do diffs in both directions, unless --no-bidirectional is passed.
We can post-hoc break the summary statistics down based on the source of the left term
[24]:
unique_srcs = df['left_source'].unique()
n_unique = len(unique_srcs)
fig, axs = plt.subplots(nrows=n_unique, figsize=(10, 6*n_unique))
for ax, left_source in zip(axs, unique_srcs):
sub_df = df[df['left_source'] == left_source]
sns.countplot(data=sub_df, x='category', hue='left_predicate_id', ax=ax)
ax.set_title(f'Count of each type broken down by left_source = {left_source}')
ax.set_xlabel('Type')
ax.set_ylabel('Count')
ax.set_yscale('log') # Make y-axis logarithmic
ax.tick_params(axis='x', rotation=45) # Make x-axis labels diagonal
plt.tight_layout() # Ensure layout is tight so labels don't get cut off
plt.show()
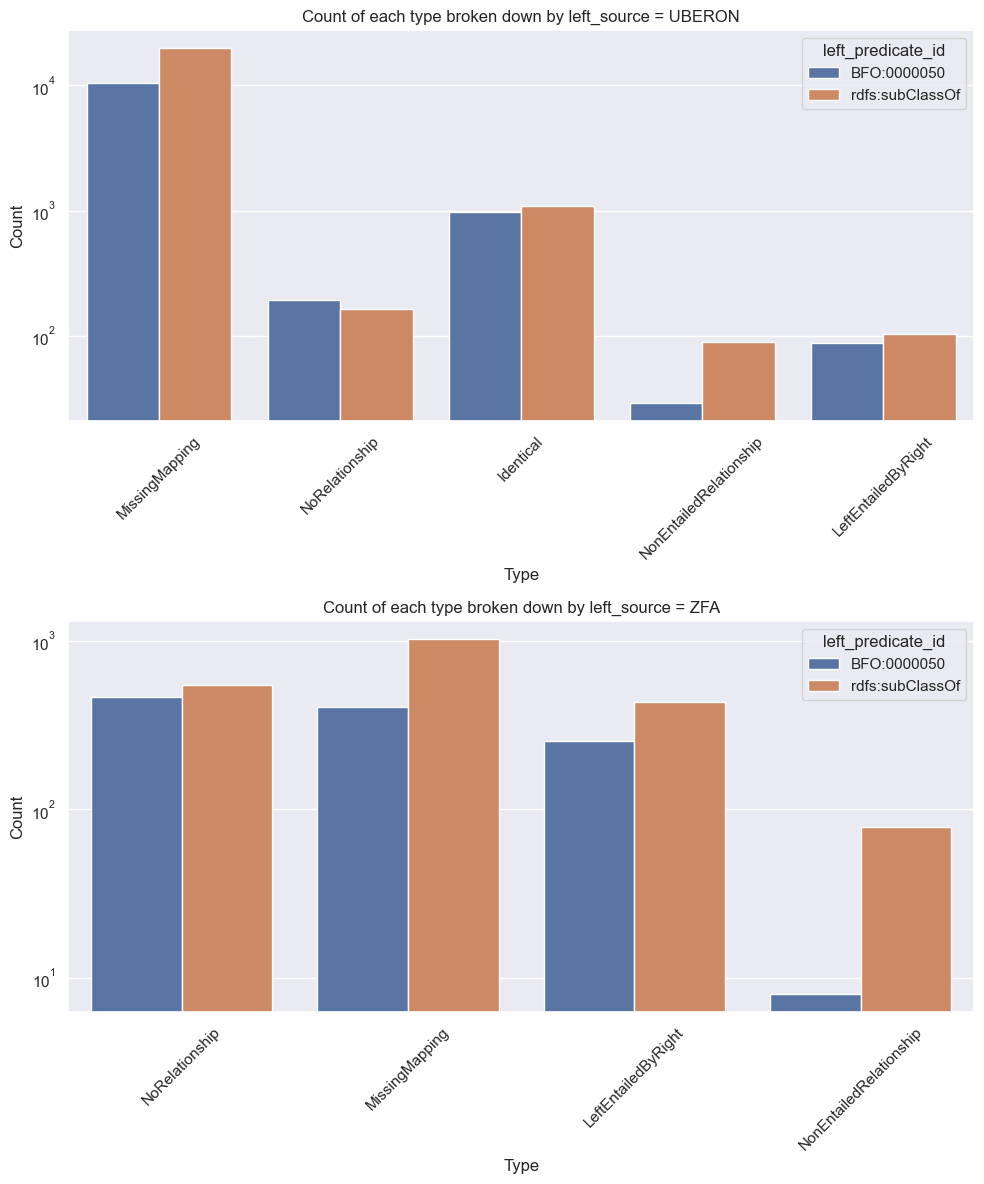
As expected MissingMapping dominates the Uberon->ZFA direction. Note this is not actually “missing” in the sense of incomplete, we expect most edges in uberon to be non-mappable to ZFA due to difference in taxonomic scope.
[ ]: